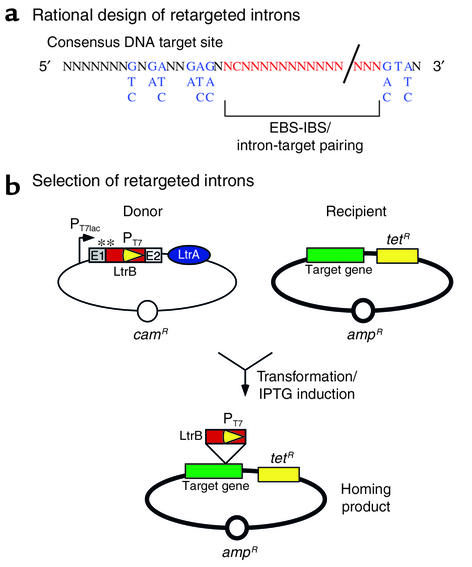
Figure 4.
Rational design and selection of retargeted introns using a mobility assay in E. coli. (a) Rational design. Putative target sites may be searched in a gene of interest using the consensus sequence shown (N is any nucleotide). At positions where a subset of nucleotides is preferred by the LtrA protein, each preferred nucleotide is listed (in blue). The EBS of the intron is designed to base pair with the predicted IBS of the target (in red). Mobility is assessed in E. coli using donor and recipient plasmids as shown in b. (b) Selection of target sites. Retrohoming into the recipient plasmid activates expression of the promoterless tetracycline resistance gene (tetR, yellow box). The donor plasmid expresses the intron (L1.LtrB, in red) and associated protein (LtrA, in blue) following transformation into E. coli and isopropyl-β-D-thiogalactopyranoside (IPTG) induction of the T7lac promoter (PT7lac). The intron is flanked by exon sequences (E1 and E2) required for self-splicing. **A library of introns is generated by randomizing EBS sequences in the intron. IBS sequences in E1 are also randomized to facilitate forward splicing of the library of precursors. A T7 promoter (yellow arrowhead) is located in domain IV of the intron. Selectable markers on donor and recipient plasmids are shown (ampR, camR). Transcription termination signals prevent TetR expression in the absence of retrohoming. Retargeted introns are recovered from ampR, tetR colonies.