FIG. 1.
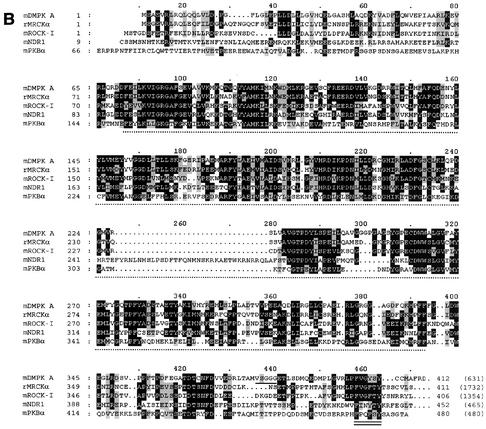
DMPK: domain organization and homology to serine/threonine kinase family members. (A) Major DMPK isoforms A to F have an N-terminal leucine-rich domain (L), a serine/threonine kinase domain, a protein kinase C-terminal domain containing the hydrophobic phosphorylation motif, and a coiled coil region. Differences between isoforms originate from alternative splicing, conserved between humans and mice: (i) a VSGGG-sequence can be present (isoforms A, C, and E) or absent (isoforms B, D, and F) and (ii) three different C-terminal tails occur. Minor splice form DMPK G, only present in humans, carries a fourth type of C terminus, of which the N-terminal half is identical to tail 1. (B) Sequence comparison between mDMPK, rMRCKα, mROCK-I, mNDR1, and mPKBα. Only the first 412 aa of DMPK are shown, since no relevant homology exists with the other kinases beyond this point. Identical amino acids (in at least three of the five kinases) are shown in white on a black background, and similar amino acids are shown in black on a grey background. The kinase domain is indicated with a dotted line below the sequence, the VSGGG sequence is underlined, and the hydrophobic phosphorylation motif is doubly underlined. The total number of amino acids for each full-length protein is indicated in parentheses; note that rMRCKα and mROCK-I are very large proteins compared to mDMPK (accession numbers: P54265, T14039, S74244, AAH09658, and P31750). (C) Sequence identity between mDMPK, rMRCKα, ROCK-I, mNDR1, and mPKBα. The N terminus (aa 1 to 70), the kinase domain (aa 71 to 339), and the protein kinase C-terminal domain (aa 340 to 405) of DMPK were compared with the corresponding parts in rMRCKα, mROCK-I, mNDR1, and mPKBα using ClustalW. The relative sequence identity for each domain is expressed as a percentage relative to mDMPK. Values for the protein kinase C-terminal domain without the VSGGG motif (as in DMPK B, D, and F) are listed in parentheses. Similar results were obtained with rMRCKβ and mROCK-II (data not shown).