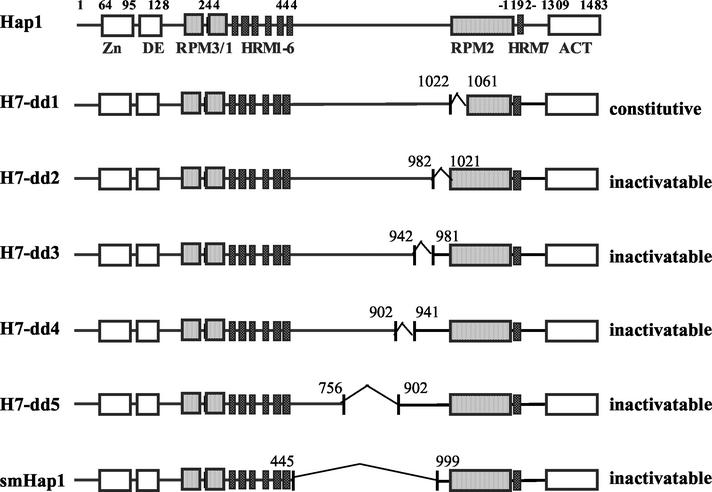
FIG. 4.
Hap1 domain structure and schematic diagrams of Hap1 mutants with deletions between HRM1 to -6 and RPM2. Shown are the C6 zinc cluster motif (Zn), the dimerization element (DE), three repression modules (RPM1 to -3), seven heme-responsive motifs (i.e., HRM1 to -6 and HRM7), and the activation domain (ACT). The deleted regions in mutants are marked. Residues 1022 to 1061, 982 to 1021, 942 to 981, 902 to 941, 756 to 902, and 445 to 999 are deleted in mutants H7-dd1, H7-dd2, H7-dd3, H7-dd4, H7-dd5, and smHap1, respectively. Mutant H7-dd1 was constitutively active—its activity in the presence of the heme analogue dpIX was as high as that in the absence of dpIX. All other mutants are inactivable—their activity was as low as the basal level activity and remained unchanged in cells grown in increasing concentrations of dpIX. Hap1 activity was measured as β-galactosidase activity expressed from the UAS1/CYC1-lacZ reporter gene.