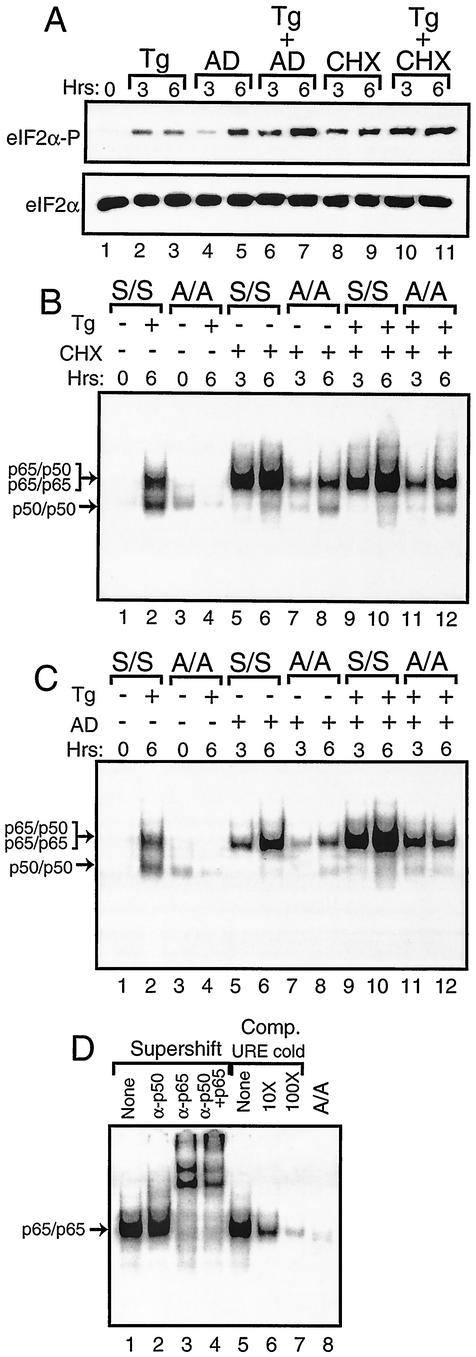
FIG. 8.
Activation of NF-κB in response to cycloheximide or actinomycin D exposure requires phosphorylation of eIF2α. (A) S/S MEF cells were exposed for the indicated number of hours to thapsigargin (Tg), actinomycin D (AD), or cycloheximide (CHX) as indicated or to no stress agent (0 h). Phosphorylation of eIF2α was measured by immunoblot analysis using polyclonal antibody specific to eIF2α phosphorylated at Ser-51 (eIF2α∼P). Levels of total eIF2α were assayed by using antibody that recognizes both phosphorylated and nonphosphorylated versions of the translation initiation factor (eIF2α). S/S and A/A MEF cells were exposed to thapsigargin, cycloheximide (B), or actinomycin D (C) as indicated by the “+” or “−” for the indicated number of hours. Nuclear lysates prepared from S/S and A/A MEF cells, as indicated, were analyzed for binding to the NF-κB probe in the EMSA. Arrows indicate DNA complexed with p65/p50 or p50/p50 in thapsigargin-stressed preparations or with p65/p65 in cycloheximide or actinomycin D-stressed cells as defined in experiments shown in panel D. In panel D, S/S MEF cells were treated with actinomycin D for 6 h and analyzed for binding with the NF-κB DNA. In lanes 1 to 4, supershift designates that antibodies that specifically recognize p50 and/or p65 were added to the EMSA binding mixture. “None” indicates that no antibody was added to the assay. In lanes 5 to 7, competition (Comp) indicates that nonradiolabeled NF-κB URE competitor DNA was added at a 10× or 100× molar excess or was not added (None). In lane 8, A/A MEF cells were similarly analyzed using actinomycin D.