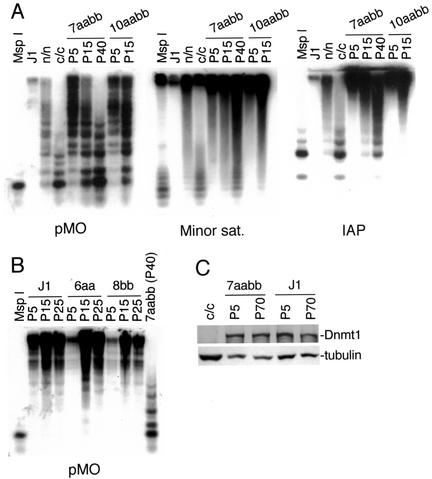
FIG. 1.
Inactivation of Dnmt3a and Dnmt3b results in progressive loss of DNA methylation in ES cells. (A) Genomic DNA from Dnmt3a−/− Dnmt3b−/− ES cells (7aabb and 10aabb) that had been grown in culture for 5 to 40 passages, as well as wild-type (J1) and Dnmt1 mutant (n/n and c/c) ES cells, was digested with HpaII and hybridized to probes for endogenous C-type retrovirus repeats (pMO), minor satellite repeats, and IAP repeats. As a control for complete digestion, DNA from J1 cells was digested with MspI. The Dnmt1n allele (the “n” stands for N-terminal disruption) is a partial loss-of-function mutation (27) and the Dnmt1c allele (the “c” stands for disruption of the catalytic or C-terminal domain) is a null mutation (24). (B) Genomic DNA from J1, Dnmt3a−/− (6aa), or Dnmt3b−/− (8bb) ES cells that had been grown in culture for 5 to 25 passages, as well as 7aabb (P40), was digested with HpaII and hybridized to the pMO probe. (C) Lysates from the indicated ES cell lines were immunoblotted with anti-Dnmt1 and anti-tubulin antibodies.