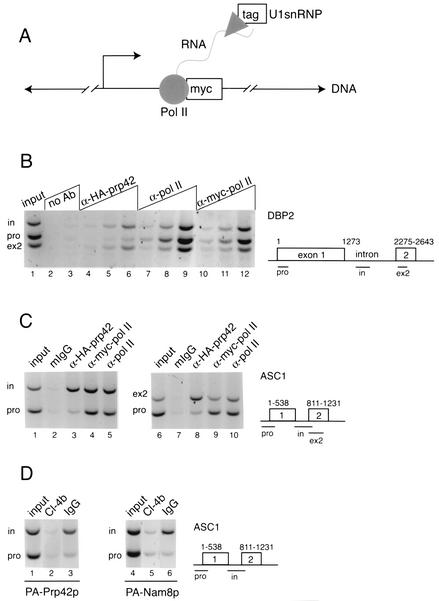
FIG. 1.
Detection of the U1 snRNP at intronic and downstream segments of DBP2 and ASC1 genes by ChIP. (A) Experimental design. In an endogenous transcription unit, the tagged U1 snRNP (Prp42p or Nam8p, triangle) is tethered to the chromatin by nascent RNA via Myc-tagged RNA Pol II (Rpo21p, circle). After formaldehyde cross-linking, random shearing of DNA (hatch marks), and specific immunoprecipitation for the tag, coimmunoprecipitated DNA will be detected by PCR. (B) PCR detection of specific regions within the DBP2 gene in the starting YKK19 extract (input, lane 1), control beads (no Ab, lanes 2 and 3), and ChIPs carried out with antibodies specific for the HA-tagged Prp42p (lanes 4 to 6), Pol II itself (8WG16, lanes 7 to 9), and Myc-tagged Pol II (lanes 10 to 12). A schematic diagram of DBP2 shows the layout of the ORF with respect to introns and exons, as well as the locations of predicted PCR products representing the promoter (pro), intron (in), and second exon (ex2) regions. A titration of ChIP templates used for PCR is shown, in which 0.3 μl (lanes 4, 7, and 10), 1.0 μl (lanes 2, 5, 8, and 11), and 3.0 μl (lanes 3, 6, 9, and 12) were added. Data shown here is representative of four independent ChIP experiments. (C) PCR detection of promoter (pro), intron (in), and exon2 (ex2) regions of the ASC1 gene in the starting extract (input, lanes 1 and 6), mouse nonimmune IgG control ChIP (mIgG, lanes 2 and 7), and ChIPs carried out with antibodies specific for the HA-tagged Prp42p (lanes 3 and 8), Myc-tagged Pol II (lanes 4 and 9), and Pol II itself (8WG16, lanes 5 and 10). A schematic diagram of ASC1 shows the layout of the ORF and the locations of predicted PCR products. The data shown here are representative of three independent ChIP experiments. In this experiment, Prp42p ChIP templates yielded intron DNA levels 4.6-fold and ex2 DNA levels 7.4-fold above promoter levels, relative to Pol II. (D) Detection of elevated U1 snRNP levels within the ASC1 intron in strains harboring protein A-tagged Prp42p or Nam8p, after ChIP with rabbit IgG-coated beads. The data shown here are representative of three (PA-Prp42p) and two (PA-Nam8p) independent ChIP experiments.