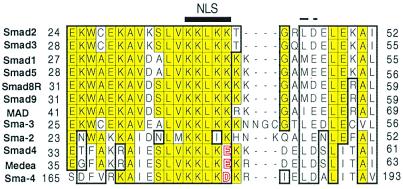
Figure 2.
Multiple sequence alignment of the presumptive NLS regions of Smad proteins. N-terminal regions from pathway-specific Smads 1, 2, 3, 5, 8, and 9 (all human except Smad 8, which is from rat), their Drosophila and C. elegans homologues (Mad, Sma-2, and Sma-3), and the co-Smad protein Smad 4 and its Drosophila and C. elegans homologues (Medea and Sma-4) were aligned with clustalx software and displayed by the seqvu program (using Smad 2 as primary sequence). Identical residues are yellow colored, and homologous residues are boxed. The identified five-residue NLS-like motif is highlighted. Smad 4 and its homologues contain an “E” or “D” in place of “K” in the last position of the NLS; these acidic residues are shaded red. The two conserved hydrophobic/acidic residues at the C-terminal side of the NLS are indicated by a dashed line.