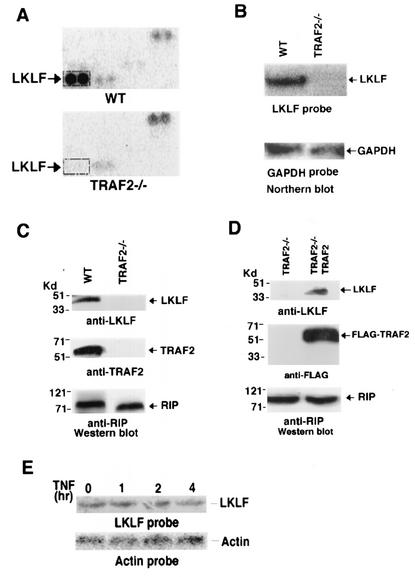
FIG. 1.
Impaired LKLF expression in TRAF2−/− cells. (A) Two microarray filters were hybridized with the cDNA probes generated from RNAs from WT and TRAF2−/− mouse fibroblast cells. The results of a portion of the microarrays are shown. The spots of LKLF cDNA are indicated. (B) Equal amounts of total RNA from WT and TRAF2−/− cells were Northern blotted for LKLF with a LKLF cDNA probe. The equal input of RNA from each sample was verified with a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe. (C) Equal amounts of cell extracts from WT and TRAF2−/− cells were Western blotted for LKLF, TRAF2, and RIP proteins. RIP was used as a protein input control. (D) Equal amounts of cell extracts from TRAF2−/− cells and TRAF2−/− cells in which LKLF expression was reconstituted by ectopically expressing TRAF2 were Western blotted for LKLF, FLAG-TRAF2, and RIP proteins. RIP was used as a protein input control. In panels C and D, the positions of molecular mass markers (in kilodaltons) are shown to the left of the blots. (E) Equal amounts of total RNA from WT fibroblasts treated with TNF (30 ng/ml) for the indicated time points were Northern blotted with LKLF and actin probes.