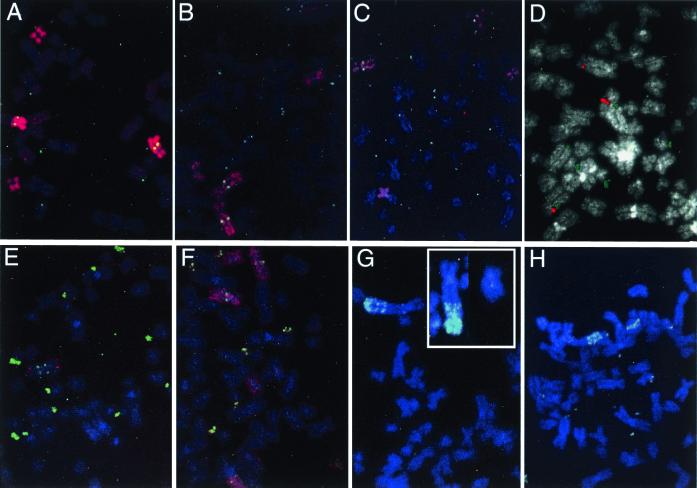
Figure 4.
FISH patterns demonstrating seven of the eight types of chromosome rearrangement detected. (A) The 5 paint/DHFR combination on A6.8/100, showing the loss of a normal 5 and the presence of several DMs. (B) The 5 paint/DHFR combination on A6.7/100, showing the loss of the der(3p5q) and the presence of numerous DMs. (C) The 5 paint/DHFR combination on A6.3/100, showing retention of two normal 5s, DMs, and a small fragment of der(3p5q) translocated to an unidentified chromosome (lower left). (D) The C5–92 cosmid (red) and DHFR (green) probe combinations on A6/100, showing the presence of two normal 5s, numerous DMs, and a third chromosome hybridizing to the C5–92 probe only. (E) A C5–9/DHFR combination on A6.5/100, showing the presence of only one normal 5, the der(3p5q) at the bottom, an i(5q), and numerous DMs. (F) A 5 paint/DHFR combination on A6.12/20 (the pattern is similar to A6.12/100), showing two normal 5s, a der(3p5q) on the right side, two i(5p)s, and numerous DMs. (G) The C5–92/DHFR combination on A6.7g, illustrating two normal chromosomes 5 and the HSR on the der(3p;inv5q); C5–92 is retained at its original position on all three chromosomes. (Inset) The C5–9/DHFR combination, showing the der(3p;inv5q) and a normal 5 from another mitotic cell, which best illustrates the inversion within the 5q arm. (H) An example from a second group of A6.7g cells that carries copies of the DHFR gene as DMs, in an HSR near the terminus of the der(3p;inv5q), and in large fragments of the HSR. The probe combination is C5–9/DHFR. Two normal 5s are present in each cell.