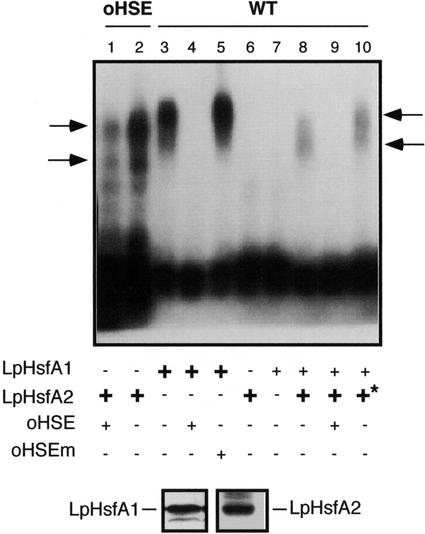
Figure 4.
In vitro binding experiments. Two labeled DNA fragments that contained synthetic (oHSE) or natural HSE sequences from Ha hsp17.6 G1 (WT) were subjected to mobility shift assays in agarose gels. Binding reactions contained the probe indicated on top (0.13 ng of labeled DNA) and the components were summarized at the bottom, including unlabeled oligonucleotide fragments used as specific or unspecific competitors (oHSE and oHSEm, respectively; see “Material and Methods”), as well as different amounts of protein extracts from yeast strains containing LpHsfA1 or LpHsfA2. Optimal and limiting amounts of total protein used for each extract were experimentally determined (see “Results”): LpHsfA1, limiting = 10 μg (+), and optimal = 20 μg (+). In the case of LpHsfA2, 40 μg (+) was used as the optimal amount of total protein. Higher amounts of this extract were used in some reactions (+*, 70 μg). Ticks to the left mark the position of the LpHsfA2 homo-oligomeric complexes. The arrows to the right mark the position of the different mixed complexes observed with increasing amounts of the LpHsfA2 extract (lane 8, 40 μg of protein, and lane 10, 70 μg of protein). Bottom, Western immunodetection of LpHsfA1 and LpHsfA2 using 25 μg of total protein from the respective extracts used in the binding reactions.