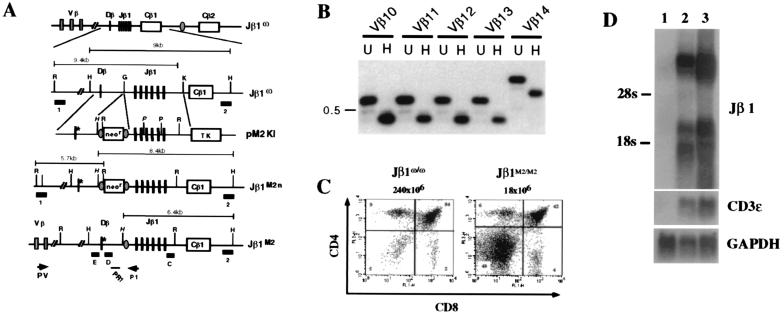
Figure 3.
Generation and analysis of mice with M2 mutation in the endogenous TCRβ locus. (A) Schematic of the Jβ1ω allele showing some of the Vβ gene segments, the DJβ1 gene cluster, the loxP site (shaded oval) that replaces the DJβ2 gene cluster, and the Cβ1 and Cβ2 genes (not to scale). Jβ1ω/ω ES cells were transfected with the pM2KI targeting vector to generate the Jβ1M2 n allele. Cre-mediated deletion of the loxP-flanked neor gene generates the Jβ1M2 allele which differs from the Jβ1ω allele by the presence of the M2 mutant heptamer (*), an introduced HindIII site (H), and a single loxP site that is not contiguous with the mutation. Shown are EcoRI (R), HindIII (H), and KpnI (K) sites. The position of probes 1, 2, C, D, and E are shown as filled rectangles. Probes 1 and 2 were used to identify correctly targeted alleles (data not shown). Also shown is the position of oligonucleotide primer P1 and probe PR1. PV is a described (20) set of Vβ gene segment primers. (B) PCR analyses of Jβ1M2/ω thymocyte DNA by using the PV panel of Vβ primers and the P1 primer. Shown are PCR reactions by using Vβ10- to Vβ14-specific primers with the P1 primer. Southern blot analysis of undigested (U) or HindIII-digested (H) PCR products was carried out by using the PR1 oligonucleotide probe. PCR products from Vβ to Dβ rearrangements on the Jβ1M2 allele are reduced in size on HindIII digestion. The 0.5-kb marker is indicated. (C) Flow cytometric analysis of thymocytes from 3- to 4-week-old Jβ1ω/ω and Jβ1M2/M2 mice by using CD4-PE and CD8-FITC. Shown is a representative analysis of the eight Jβ1ω/ω and Jβ1M2/M2 mice analyzed. (D) Whole-cell RNA was isolated from RAG-2−/− spleen (lane 1), RAG-2−/− thymus (lane 2), and Jβ1M2/M2:RAG-2−/− thymus (lane 3) and subjected to Northern blot analysis by using a probe that spans the Jβ1 gene segments (Jβ1) and the CD3ɛ and glyceraldehyde-3-phosphate dehydrogenase probes as controls.