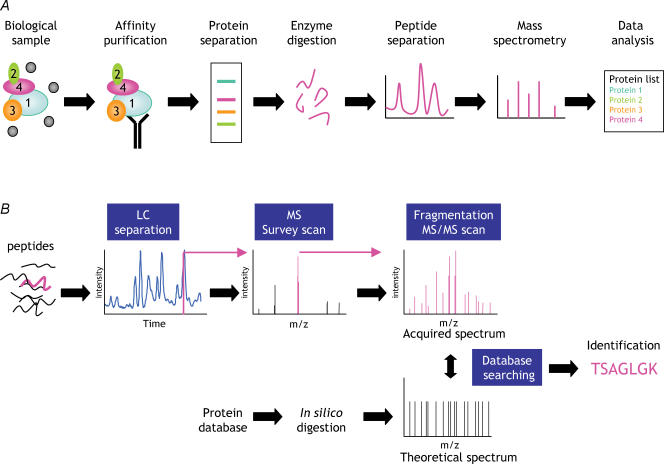
Figure 1. General strategy for protein complex identification using mass spectrometry.
A, a biological sample is purified and separated into its constituents, which are then proteolysed and analysed by LC-MS (see text for details). B, mass-spectrometry-based protein identification. A mixture of peptides (the peptide of interest is highlighted in pink) is separated by reversed-phase HPLC. The chromatography column is located immediately in-line with the MS, and peptides are analysed as they elute from the column. The mass/charge ratios (m/z) of all co-eluting peptides are first analysed in a survey (or MS) scan. Individual peptide populations are then selected (usually based on abundance) for fragmentation. Finally, the m/z of the peptide fragments are analysed to generate an MS/MS or CID spectrum. The acquired MS/MS spectrum is compared with theoretical spectra obtained via an in silico digest of a relevant protein database. Significant matches are reported, yielding peptide identification.