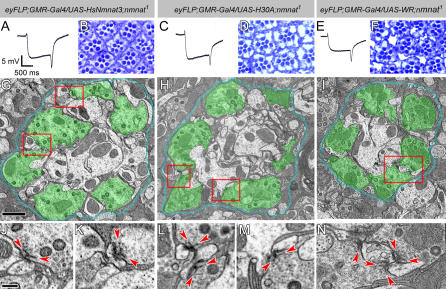
Figure 8. Enzymatically Inactive NMNAT Can Rescue the Neurodegeneration Phenotype Caused by Loss of nmnat .
(A–F) ERG recordings of mutant photoreceptors overexpressing human NMNAT3 (A), or inactive Drosophila NMNAT forms H30A (C) or WR (E) in nmnat mutant photoreceptors. The genotypes are marked on top of each column. Note that the magnitudes of both depolarization and on/off transients are partially restored. (B), (D), and (F) Retinal structures are partially restored in each genotype.
(G–I) TEM micrographs of lamina cartridges. Photoreceptor terminals are well organized in cartridges, similar to wild type. Demarcating glia are colored blue and photoreceptor terminals are colored in green to identify the structures. The red boxes in (G) indicate the regions shown in (J) and (K); the boxes in (H) indicate the regions shown in (L) and (M); and the box in (I) indicates the region shown in (N). Scale bar in (G) for (G–I) indicates 1 μm.
(J–N) Individual terminals boxed in (G–I). All active zones have defined platform (arrowheads) and pedestal structures. Scale bar in (J) for (J–N) indicates 200 nm.