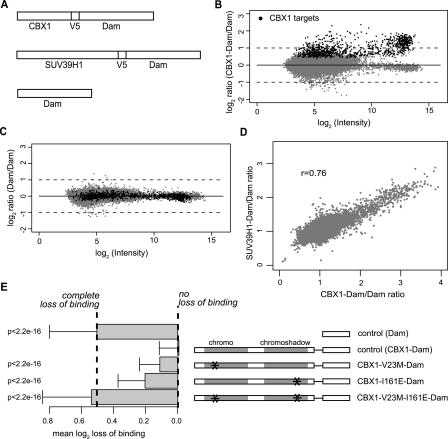
Figure 1.
CBX1 and SUV39H1 binding profiles. (A) Schematic drawing of CBX1-Dam-fusion protein, SUV39H1-Dam-fusion protein, and the Dam protein constructs that are used to generate the binding profiles. To facilitate detection of the Dam-fusion proteins a V5 epitope tag was added. Both CBX1 and SUV39H1 as well as control DamID profiles consist of the combined data of four independent experiments hybridized on four oligonucleotide arrays. (B) CBX1 DamID binding profile. For each probed sequence, the average log2 CBX1-Dam/Dam ratio is plotted against the average fluorescence intensity (log2 √(Cy5 × Cy3). Statistically significant CBX1 target loci (n = 933) are indicated by black dots. Gray dots represent nontarget loci. (C) Control DamID profile. Black dots represent CBX1 target loci projected on control data. Gray dots represent nontarget loci. (D) Bivariate scatter plot of CBX1 and SUV39H1 binding data. (r) Pearson correlation coefficient. (E) Molecular mechanism of CBX1 targeting. Overview of CBX1 mutants (right side) and bar plot showing their average loss of binding to CBX1 target loci (left side). Error bars indicate standard deviations. P-values compared to control (CBX1-wt) according to Wilcoxon rank sum test. The values representing “complete loss of binding” and “no loss of binding” were inferred from the Dam and CBX1-Dam controls, respectively. DamID profiles of CBX1-mutants consist, for each protein, of data of two independent experiments. For details on the mutants and controls, see text.