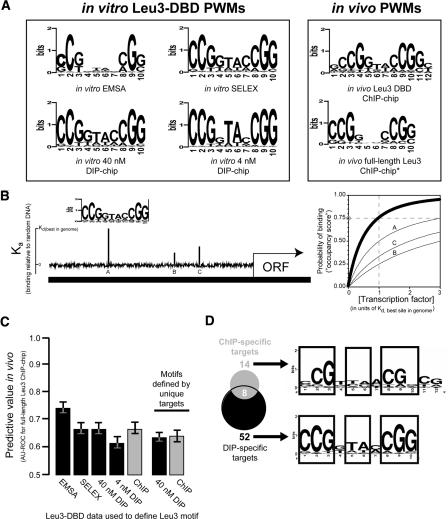
Figure 2.
Motifs derived from in vitro methods predict in vivo protein–DNA interactions as accurately as motifs derived from in vivo ChIP-chip. (A) Six PWM representations of the Leu3 binding motif, derived from the indicated binding experiments (Methods). (B) A schematic representation of motif scoring by GOMER. Briefly, given a PWM for a binding motif N bp long, GOMER calculates a relative equilibrium binding constant (Kd) for each sequence window of length N in the genome, and from this Kd value calculates the probability of being bound at some free protein concentration (typically equal to the Kd of the best site in the genome). GOMER then uses these individual binding probabilities to calculate the probability of binding to at least one site within a genomic sequence of interest. The graph (right) indicates the probability that sites A, B, and C (left) will be occupied by a factor recognizing the motif shown, as a function of protein concentration. The thick line shows the probability that any one of the three sites will be bound at the given concentration. In this example, if the protein is present at a concentration equal to the Kd of the best site in the genome, there is a 75% chance that the shown promoter will be bound at either A, B, or C at a given point in time (gray circle). (C) AUC-ROC values (y-axis) for prediction of full-length Leu3 ChIP-chip results based on motifs derived from the indicated data set. Error bars indicate the 95% confidence interval estimated using bootstrap resampling of occupancy scores and Leu3 enrichments. (D) Similar motifs are derived from genomic targets unique to DIP or ChIP.