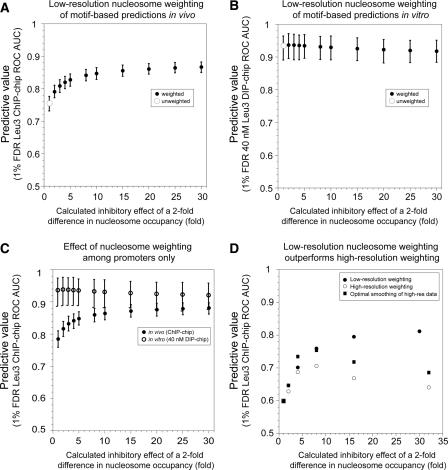
Figure 3.
Accounting for nucleosome occupancy improves target prediction in vivo but not in vitro. Improvement of in vivo Leu3–DNA interaction prediction assuming an inhibitory effect of nucleosome occupancy. All Leu3 occupancy scores were calculated using the EMSA-derived PWM. (A) Different weighting factors are shown on the x-axis. Error bars indicate 95% confidence intervals calculated by bootstrap resampling (Methods). (B) For in vitro 40 nM DIP-chip data, weighting does not significantly improve the AUC-ROC value. (C) Same as A, but excluding ORFs and intergenic sequences that lie downstream from two convergently transcribed genes. The effects of weighting on full-length Leu3 ChIP-chip experiments (solid circles) and Leu3-DBD DIP-chip (open circles) are plotted. (D) Higher-resolution nucleosome occupancy data (Yuan et al. 2005) do not offer improvement in predictions over that achieved by low-resolution data. High-resolution data are restricted to chromosome III. Weighting with low-resolution data as in panel A yields a strong improvement in predictive power (black). However, higher-resolution data do not perform as well (open circles). The high-resolution data were most predictive when computationally “blurred” over 300 bp (squares).