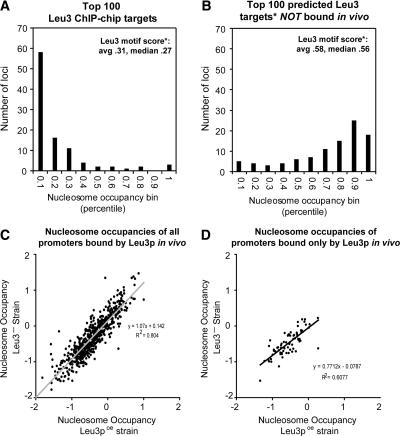
Figure 4.
Leu3-bound motifs are nucleosome-poor, but low nucleosome occupancy is not a consequence of Leu3 binding. (A) The 100 loci most highly enriched in Leu3 ChIP-chip experiments (by SAEM P-value) were divided into 10 bins according to their nucleosome occupancies relative to all other loci, as measured in a wild-type strain. The number of loci in each bin is shown on the y-axis. “Leu3 motif score” refers to the GOMER score of the arrayed locus (for all arrayed loci, the average was 0.09, median 0.06). (B) Same as A, except the 100 loci that had the highest predicted affinity to Leu3 and were not bound in vivo were plotted. Leu3 binding affinities were predicted using GOMER. (C) Nucleosome occupancy in strains overexpressing a gene encoding the Leu3 activation domain but no Leu3 binding domain (Leu3-, log2 ratios; y-axis) was highly correlated with nucleosome occupancy in strains overexpressing full-length Leu3 protein (Leu3poe, log2 ratios; x-axis). Thus, low nucleosome occupancy at Leu3-bound promoters is not dependent on Leu3 binding. The positive y-intercept and the slope slightly greater than 1 suggest there may be a subtle effect of Leu3 on nucleosome occupancy, but as shown in Figure 5, nucleosome occupancies determined in the absence of Leu3 are just as predictive of Leu3 binding as nucleosome occupancies determined in the presence of Leu3. (D) Same as C, but for the 76 Leu3 targets not bound by any other transcription factor (Lee et al. 2002; Harbison et al. 2004).