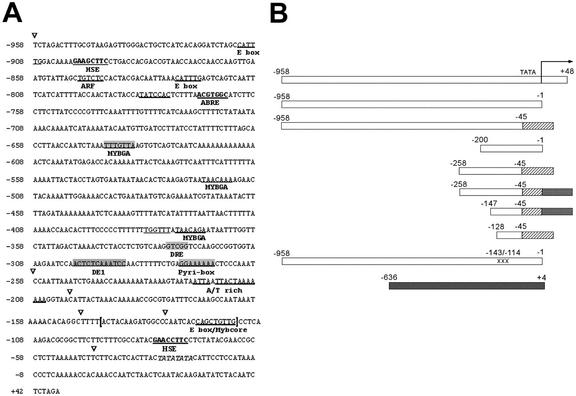
Figure 1.
5′ TCH4 genomic sequence and schematic representation of TCH4 regions generated and tested for transcriptional regulatory activity. A, Genomic sequence of TCH4 upstream region sufficient to confer up-regulation of expression. The inverted triangles indicate the 5′ positions of TCH4 regions generated and tested for transcriptional regulatory activity (see B). Sequences related to defined regulatory motifs in the sense orientation are underlined, whereas inverted motifs are shaded in gray (motifs are defined in text). The putative TATA box is indicated in italics. Numbering refers to distance from transcriptional start site designated as +1. B, White rectangles represent TCH4 sequences between −958 and +48 and subregions fused to the GUS and LUC reporter genes. The numbering refers to distance relative to the transcriptional start site (+1). The arrow indicates the start position and direction of transcription. For some constructs, the endogenous promoter was replaced with −90- or −46-bp cauliflower mosaic virus (CaMV) 35S promoter (hatched, size indicated by rectangle length); some constructs also have a 143-bp region of a 5′-UTR from tobacco etch virus (TEV) (cross hatched). The penultimate rectangle from the bottom represents a construct containing TCH4 sequences from −958 to −1 that has substituted nucleotides between −143 and −114. The black rectangle at the bottom represents the sequences present from the UBQ10 locus for UBQ10::LUC. These representations of transgenes are not drawn to scale.