Abstract
Site-directed mutagenesis was used to identify cis-acting elements that control hormonal and abscission-specific expression of the bean (Phaseolus vulgaris) abscission cellulase (BAC) promoter. Auxin inhibition of BAC promoter expression is at least in part controlled by a negatively regulated element and ethylene induction by a positively regulated element. One of a series of 15 different 10-bp mutations created in a 2.9-kb BAC promoter reduced reporter gene expression by 60%. The native sequence for this 10-bp mutation includes a TGA-type basic leucine zipper (bZIP) motif. Tandem ligation of three 18-bp BAC elements (Z-BAC), which includes the bZIP motif to a minimal −50 35S cauliflower mosaic virus promoter, enhanced expression in abscission zones (AZs) 13-fold over that of the minimal promoter alone. The native forward orientation of the Z-BAC elements was essential for high expression levels. Expression of the Z-BAC minimal construct was 3-fold greater in AZ than stems when compared with the expression levels of an internal control with an enhanced 35S cauliflower mosaic virus promoter. Polymerase chain reaction was used to identify three TGA-type bZIP transcription factors in an AZ cDNA library. One of these factors was of the class I type and two of the class II type. RNA-blot analysis was completed for these genes and electrophoretic mobility shift assays used to confirm their binding to the Z-BAC element. Electrophoretic mobility shift assay-binding affinity was greatest for the class I TGA-type bZIP factor. The results indicate a complex interaction of negative and positive regulating transcription factors that control BAC gene expression.
Abscission (organ separation) is a process common to higher plants. The shedding of plant parts, both reproductive and vegetative, is important for reproduction, plant defense, resistance to drought and flooding, and continuation of perennial growth (Sexton and Roberts, 1982). Abscission occurs by degradation of the primary cell wall and middle lamella surrounding cells in a separation layer that forms within a broad region of cells commonly referred to as the abscission zone (AZ). One of the first model systems used for abscission research was bean (Phaseolus vulgaris). It was in bean leaf AZs that the first abscission-specific hydrolase, β-1,4-endo-glucanase, was characterized (Lewis and Varner, 1970), purified (Koehler et al., 1980), and the cDNA (Tucker et al., 1988) and genomic (Koehler et al., 1996) clones identified. Although the control of abscission is not identical for all plant parts, a common pattern for the regulation of abscission is that ethylene induces and enhances the process, whereas auxin strongly inhibits it (Sexton and Roberts, 1982). Expression of the bean abscission cellulase (BAC) mRNA correlates precisely with the onset of abscission and its regulation by ethylene and auxin (Tucker et al., 1988). Moreover, after ethylene treatment to induce abscission, expression of the BAC mRNA is limited to a depth of one or two cells on either side of the fracture plane in nonvascular tissue and up to 3 mm distal to the fracture plane in cells within the vascular bundle. All cell types in the separation layer and vascular bundle accumulate BAC mRNA (Tucker et al., 1991).
Currently, an understanding of the regulatory mechanisms that control hormonal and tissue-specific expression of abscission-specific genes is limited. To further enhance our understanding of these processes, we embarked on a study of the BAC promoter to identify cis-elements and trans-acting factors that regulate abscission-specific expression. Earlier studies with the BAC promoter in stably transformed tomato (Lycopersicon esculentum) and transient expression assays in bean explants demonstrated that 210 bp of the proximal 5′-upstream BAC sequence was sufficient for ethylene- and auxin-regulated expression (Koehler et al., 1996). In bean explants, expression from this minimal BAC promoter was restricted primarily to the AZ. In addition, comparison of the BAC promoter sequence with that of its isologue in soybean (Glycine max) revealed high sequence identity within the proximal region of the two gene promoters.
Here, we describe specific elements within the proximal BAC promoter (−23 to −172) that govern the hormonal regulation of a 2.9-kb BAC gene promoter and also control its tissue-specific expression. The most notable positive regulatory element within the proximal promoter includes a TGA-type basic Leu zipper (bZIP) DNA-binding motif. The regulation of this cis-acting element was characterized in greater detail and its binding to TGA-type bZIP transcription factors isolated from AZ analyzed by an electrophoretic mobility shift assay (EMSA).
RESULTS
Site-Directed Mutagenesis of the Proximal Region of the BAC Promoter
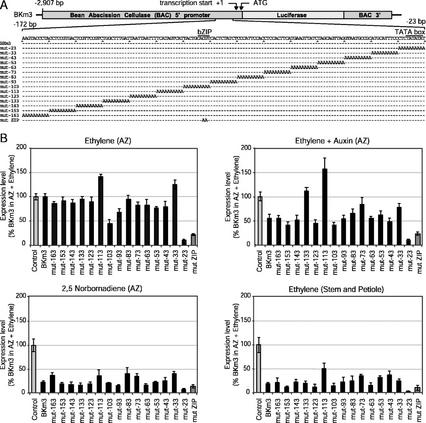
Earlier studies demonstrated that a 210-bp proximal BAC promoter retained tissue specificity and hormonal regulation similar to a 2.9-kb BAC promoter, but at a much reduced level of expression (Koehler et al., 1996). To characterize specific elements within the proximal BAC promoter, site-directed mutagenesis was used to generate a series of 15 substitutions of 10 bp each between −172 and −23 bp (relative to the start of transcription) in a full-length 2.9-kb BAC promoter (BKm3; Fig. 1A). The full-length promoter was used to maintain a high level of expression in the transient expression assay that was easily and accurately measured. Although the bean explants synthesize ethylene during the 48-h incubation, ethylene was added to the incubation chamber to synchronize the induction of abscission and BAC gene expression in the bombarded explants. Most of the 10-bp substitutions in the proximal region of the BAC promoter inhibited reporter gene expression in ethylene-treated AZ by 25% or less (Fig. 1B). However, expression levels for mutants −23, −93, and −103 were more significantly reduced by 90%, 30%, and 60%, respectively. Mutant −23, which had 90% less expression, included the TATA box. In contrast, mutant −33 immediately upstream from the TATA box, displayed a 20% increase in expression, and mutant −113 displayed a 40% increase in expression in ethylene-treated AZ. Auxin (10−4 m 2,4-D), which strongly inhibits abscission and cellulase gene expression even in the presence of high concentrations of ethylene (Tucker et al., 1988; Koehler et al., 1996), inhibited expression from the native BKm3 and most of the mutant constructs by 40% to 50% (Fig. 1B). However, expression levels for those mutants that were already reduced in ethylene-treated AZ, the TATA box mutant and mutants −93 and −103, were not significantly reduced further by the auxin treatment. Interestingly, mutants −73, −113, and −133, which displayed 83%, 140%, and 95% expression, respectively, in ethylene-treated AZ, showed no reduction of expression in response to auxin and remained at the same high levels as observed in treatments with ethylene alone. NBD is a competitive inhibitor of ethylene action (Sisler et al., 1985). Because ethylene is endogenously synthesized by the explants, NBD must be added to fully suppress ethylene responses. Addition of the gaseous NBD to the incubation chamber suppressed expression of the unmutated BKm3 construct and all the mutant constructs (Fig. 1B). Expression levels in ethylene-treated stems and petioles were similar to that in AZ treated with NBD (Fig. 1B).
Figure 1.
Construction of 10 bp poly(A+) substitutions and bZIP mutation in the 2.9-kb BAC promoter (BKm3) and their transient expression in bean explants. A, Construct design and graphical representation of substitutions within the proximal BAC promoter region. B, Transient expression results. Expression levels are the ratios of luciferase to GUS activity normalized to the ratio for the external control that consists of a native BAC promoter in BKm3 shot into AZ explants exposed to ethylene for 48 h. Treatments were 1 μL L−1 ethylene in air, auxin as 10−4 m 2,4-dichlorophenoxyacetic acid (2,4-D), and 5,000 μL L−1 2,5-norbornadiene (NBD). Each mean and se represents six or more replicate plates containing nine AZ explants or five stems and 10 petiole explants.
The 10-bp mutation (mut −103) involved in positive regulation of the BAC promoter contains an ACGT core motif recognized as a binding site for bZIP transcription factors (Izawa et al., 1993). Mutation of the ACGT motif to AaaT reduced ethylene-induced expression of the 2.9-kb BAC promoter by 80% (Fig. 1B). Expression remained low in AZ treated with auxin or NBD and in ethylene-treated stems and petioles (Fig. 1B). These results suggest that the bZIP element plays a primary positive role in the regulation of BAC gene expression.
Independence of the Element Including the bZIP Motif
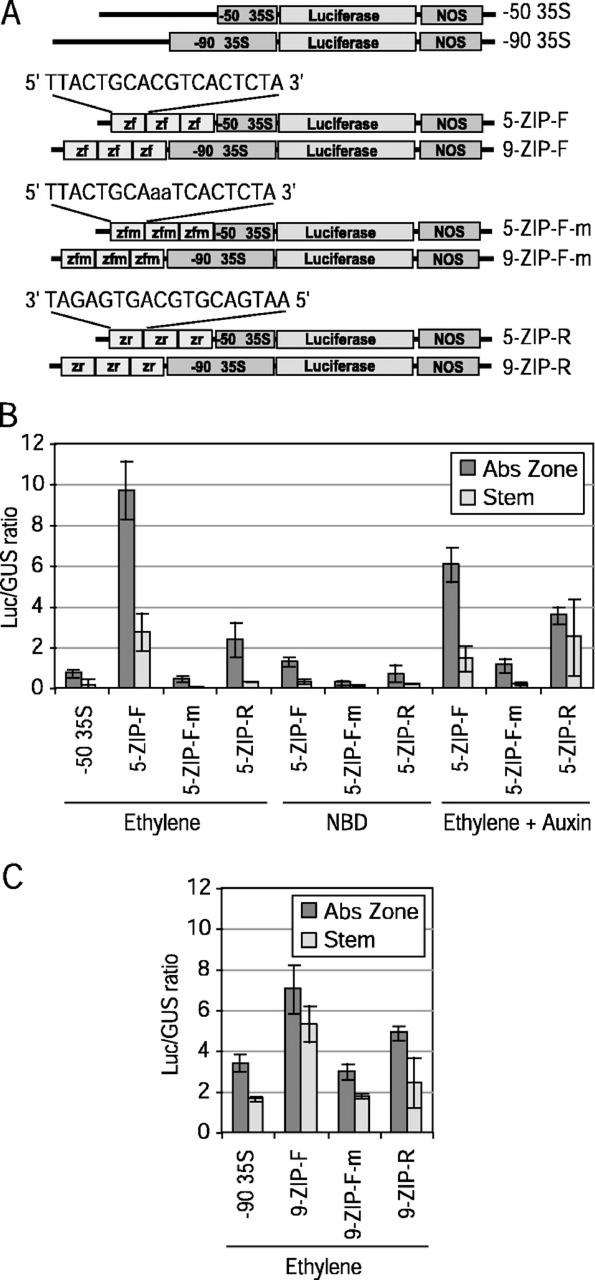
To determine the independence of the bZIP-containing element in the regulation of BAC gene expression, constructs were prepared that included this region fused to minimal cauliflower mosaic virus (CaMV) 35S promoters of two different lengths (−50 and −90; Fig. 2A). The BAC element used in these constructs (Z-BAC) spanned the region between −96 and −113. This element includes the ACGT motif at its center and seven nucleotides on either side of it (18 bp total). Three Z-BAC elements were fused in tandem to both the −50 and −90 minimal promoters (5-ZIP-F and 9-ZIP-F, respectively; Fig. 2A). In addition to the forward, native orientation of the Z-BAC element, constructs were prepared with the tandem elements in the reverse-complement orientation (5-ZIP-R and 9-ZIP-R). Furthermore, similar forward orientation constructs were prepared where the core bZIP ACGT motif was mutated to AaaT (5-ZIP-F-m and 9-ZIP-F-m).
Figure 2.
Transient expression characteristics of two minimal CaMV 35S promoters (−50 and −90) fused to three tandem 18-bp repeats of the native, mutant, or reverse complement of the Z-BAC element. A, Luciferase constructs with a minimal −50 and −90 35S promoter fused to tandem repeats of the native Z-BAC element (zf), mutant Z-BAC element (zfm), and reverse complement Z-BAC element (zr). Relative transient expression of the −50 35S (B) and −90 35S (C) minimal constructs in bean AZ and stem explants. Ethylene, auxin, and NBD treatments are as described in Figure 1. Unmodified ratios for luciferase to GUS activity are shown to allow a direct comparison of the expression levels from the −50 and −90 35S minimal constructs. Each mean and se represents four replicate plates containing nine AZs or nine stem explants.
The −50 CaMV 35S construct without addition of any BAC sequence (−50 35S) and the three different BAC fusions (5-ZIP-F, 5-ZIP-F-m, and 5-ZIP-R) were used for transient expression assay (Fig. 2B). In these experiments, the e35S::GUS construct was used as an internal control. Addition of the three tandem Z-BAC elements to the −50 35S promoter enhanced expression in ethylene-treated AZ 13-fold (Fig. 2B). Expression was also enhanced in ethylene-treated stems, but the absolute level of expression was only 35% of that in AZ. Mutation of the bZIP core motif in the three tandem repeats reduced expression levels to that of the −50 35S construct with no additional BAC sequence added. Moreover, the reverse orientation of the Z-BAC element was approximately 20% of the forward orientation, suggesting that the orientation of the Z-BAC element is important to its regulation. Exposure to NBD almost completely eliminated the enhancement supported by the Z-BAC sequence. Auxin inhibited expression by approximately 40% in ethylene-treated AZ and stem tissues that were bombarded with the construct containing the wild-type Z-BAC element (Fig. 2B). In addition, auxin appeared to stimulate expression in AZ and stems bombarded with the wild-type Z-BAC element in reverse orientation (Fig. 2B).
Expression of the −90 35S construct without additional BAC sequence was approximately 2-fold higher in AZ than stem. Addition of the three tandem Z-BAC elements further enhanced expression of the −90 35S construct by only 2-fold in both the AZ and stems. In addition, when the ACGT motif was mutated to AaaT, expression was essentially the same as the −90 35S construct, which has no BAC sequence. The reverse complement of the Z-BAC elements in the −90 35S constructs was also less effective than the forward native orientation (Fig. 2C).
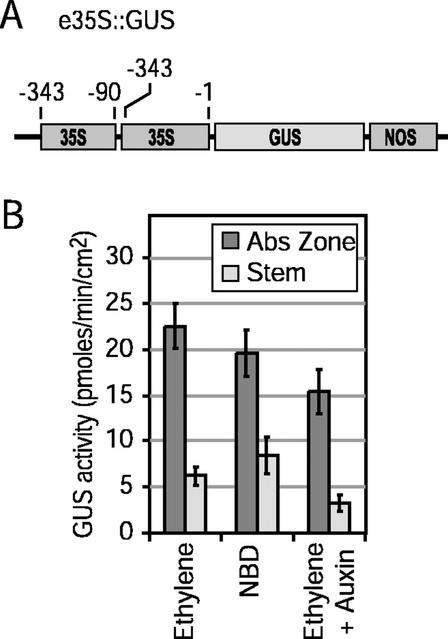
Expression Characteristics of the e35S::GUS Internal Control
An internal control was used to normalize transient expression data and reduce variability among replicates. An enhanced CaMV 35S promoter (Kay et al., 1987) was used for this purpose. The data displayed in Figure 3B are the means for e35S::GUS expression of all the replicates completed for the minimal promoter studies shown in Figure 2. The means and ses represent 30, 20, and 18 replicate bombardments for AZ and stems exposed to ethylene, NBD, or ethylene and auxin, respectively (Fig. 3B). The GUS assays were done on a per gram fresh weight bases; however, because only the surface 1 mm of AZ and stems was harvested for assay, the weight of the tissue is directly proportional to the surface area that was bombarded. Overall, the expression of the e35S::GUS construct, independent of the treatment, was approximately 3-fold higher in AZ than stems, i.e. 3.6, 2.3, and 4.8 in ethylene, NBD, or ethylene and auxin, respectively (Fig. 3B). Treatments including NBD or auxin reduced e35S::GUS expression in AZ by 13% and 32%, respectively (Fig. 3B).
Figure 3.
Transient expression characteristics of the e35S::GUS construct in AZs and stems. A, Construct design. B, Absolute expression levels of GUS activity in AZs and stems for all the replicates used in the normalized data displayed in Figure 2. Each mean and se represents 30, 20, and 18 replicate plates exposed to ethylene, NBD, or ethylene plus auxin, respectively, as described in Figure 2.
Isolation of cDNAs Encoding TGA-Type bZIP Transcription Factors
Degenerate oligonucleotides were designed to anneal within the highly conserved basic domain of TGA-type bZIP gene sequences. Three different PCR products encoding TGA-type bZIP proteins were amplified in total RNA from ethylene-induced bean leaf AZ. Subsequently, using the PCR products as probes, the corresponding full-length cDNAs were isolated by conventional screening of a bean AZ cDNA library (Table I). In accordance with the classification for TGA-type bZIP transcription factors (Niggeweg et al., 2000b), which is based on protein sequence identities in the C-terminal region of the proteins, PvTGA1.1 aligns with class I TGA-type bZIP factors (Katagiri et al., 1989; Schindler et al., 1992), whereas PvTGA2.1 and PvTGA2.2 align with factors in the class II category (Zhang et al., 1993; Niggeweg and Gatz, 1997; Table II).
Table I.
Sequence information for transcription factors isolated from bean leaf AZ
| Clone | Accession No. | Length | Closest Match in GenBank and Percent Amino Acid Identity |
|---|---|---|---|
| bp | |||
| PvTGA1.1 | AF402607 | 1,589 | Soybean TGACG-binding (STGA1); (L28005); 95% |
| PvTGA2.1 | AF402608 | 1,940 | Vicia faba DNA-binding protein (M81827); 84% |
| PvTGA2.2 | AF402609 | 2,528 | Tobacco (Nicotiana tabacum) bZIP (TGA2.2); (AF031487); 84% |
Table II.
Percent identities of the C-terminal region of PvTGA1.1, PvTGA2.1, and PvTGA2.2 with class I and class II TGA-type bZIP transcription factors from Arabidopsis and tobacco
| Clone | Class I NtTGA1a | Class I AtTGA1 | Class II NtTGA2.1 | Class II AtOBF5 |
|---|---|---|---|---|
| % | ||||
| PvTGA1.1 | 67 | 73 | 60 | 55 |
| PvTGA2.1 | 56 | 56 | 80 | 79 |
| PvTGA2.2 | 58 | 55 | 80 | 75 |
DNA-Binding Studies
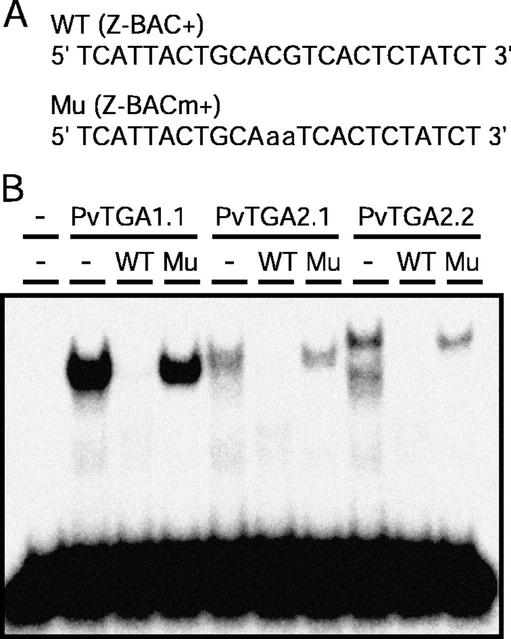
EMSAs were performed to further assess the ability of the transcription factors to bind the Z-BAC promoter sequence. The probe (Z-BAC+) consisted of the 4-bp ACGT motif at its center plus an additional 10 nucleotides of BAC sequence on either side of the motif (Fig. 4A). A coupled in vitro transcription/translation reaction using T7 RNA polymerase and rabbit reticulocyte lysate was used to express the coding sequences of PvTGA1.1, PvTGA2.1, and PvTGA2.2. Single DNA-protein complexes of various sizes were formed after incubation of PvTGA1.1, PvTGA2.1, and PvTGA2.2 proteins with the labeled Z-BAC+ probe (Fig. 4B). Because similar quantities of in vitro-synthesized PvTGA proteins were added to each of the reactions, the strong signal (5-fold) for the PvTGA1.1 complex relative to PvTGA2.1 and PvTGA2.2 complexes suggests that under the applied conditions, PvTGA1.1 had a greater affinity for the Z-BAC+ promoter sequence than either of the other two proteins. The ability of the wild-type unlabeled Z-BAC+ oligonucleotide to compete with the labeled Z-BAC+ probe, together with the inability of the unlabeled mutated probe to compete, confirms that complex formation is dependent specifically upon the CG nucleotides in the ACGT motif (Fig. 5B).
Figure 4.
EMSA for binding of bean AZ transcription factors to a 24-bp Z-BAC+ element. A, Sequence for wild-type (WT) and mutant (Mu) oligonucleotides. B, Transcription factors tested were in vitro transcribed and translated from three full-length TGA-type bZIP cDNAs isolated from a bean AZ cDNA library and competition experiments included 100-fold excess of unlabeled wild-type (WT) Z-BAC+ element or mutant Z-BAC+ element (Mu) or no unlabeled DNA added (−). First lane, Incubation where no protein or competitive DNA was added.
Figure 5.
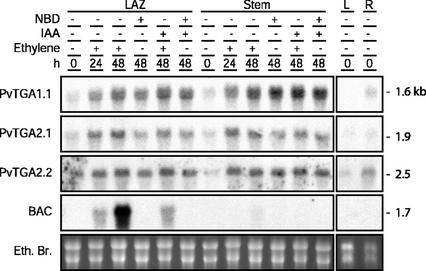
RNA-blot analysis for the expression patterns of three bean TGA-type bZIP transcription factors and the BAC gene. Several replica gels and blots were prepared from the same batch of RNA and hybridized separately with labeled cDNA probes. Leaf AZ (LAZ) or stem explants were collected and exposed to 25 μL L−1 ethylene, 10−5 m indole-3-acetic acid (IAA), and/or 5,000 μL L−1 NBD for 0, 24, or 48 h. Leaf (L) and root (R) RNA were collected from fresh untreated organs (0 h).
Expression Analysis during Ethylene- Promoted Abscission
Expression of mRNAs for each of the TGA-type bZIP transcription factors was assessed by RNA-blot analysis (Fig. 5). Transcript accumulation was analyzed in total RNA extracted from bean leaf AZ and stems (non-AZ) dissected from explants before (0 h) and after 24 and 48 h incubation in 25 μL L−1 ethylene. In addition, auxin (IAA) and NBD, an ethylene action inhibitor (Sisler et al., 1985), were applied to explants individually and in combination to determine their effect on transcription. The temporal expression patterns of the bean TGA-type bZIP mRNAs were very similar in that all three transcripts were present at low levels in both AZ and stem before ethylene exposure (Fig. 5). After incubation in ethylene for 24 or 48 h, expression of the TGA factors was enhanced to a similar extent in both AZ and stem RNA. However, NBD did not inhibit the increase in PvTGA1.1 and PvTGA2.2 transcript and only slightly reduced the increase in PvTGA2.1 transcript (Fig. 5). This suggests that the enhancement after 24 and 48 h in ethylene was not dependent on ethylene but some other cue, e.g. wounding. In addition to being independent of ethylene, IAA also had no effect on PvTGA factor expression (Fig. 5). All three TGA-type bZIP factors were expressed more abundantly in roots than leaves. PvTGA1.1 transcript was undetected in the leaves, whereas transcript for PvTGA2.1 and PvTGA2.2 were present at low levels (Fig. 5).
DISCUSSION
Earlier studies demonstrated that a 2.9-kb BAC promoter retained both the tissue-specific and hormonally regulated properties of the native gene (Koehler et al., 1996). Moreover, a much shorter 210-bp proximal promoter also retained tissue specificity and hormonal regulation but at a much reduced level of expression. Here, we confirmed these findings and extended them to demonstrate that the proximal 150 bp of the BAC promoter contains both positive and negative cis-acting elements. More importantly, it appears that the auxin and ethylene regulation of BAC gene expression may be partly independent of each other (Fig. 1B). Of particular interest are the two adjacent 10-bp mutated elements between −103 and −123 (Fig. 1B). One mutant (mut −103) is a positive regulatory element that appears to be essential for tissue-specific and ethylene-regulated BAC gene expression. This 10-bp element contains an ACGT core motif at its proximal end that is recognized as a binding site for bZIP transcription factors (Izawa et al., 1993). Mutation of the ACGT motif to AaaT reduced ethylene-induced expression of the 2.9-kb BAC promoter in AZ by 80% (Fig. 1B). The other immediately upstream 10-bp element, mut −113, is negatively regulated. Mutation of this 10-bp element enhanced BAC promoter expression and made the BAC promoter unresponsive to auxin (Fig. 1B). A more detailed mutational analysis might show that the negative regulatory element in mut −113 partly overlaps with the positive regulation in mut −103 (Fig. 1B). This possibility is supported by the observation that a simple 2-bp mutation (mut ZIP) within the ACGT motif had a greater inhibitory effect on BAC promoter expression than the 10-bp mutation that included the ACGT motif at its proximal end (Fig. 1B).
A regulatory role for the region that includes the bZIP motif (Z-BAC element) was further characterized by fusing three tandem 18-bp Z-BAC elements, with the ACGT motif at each elements center, to a −50 and −90 minimal CaMV 35S promoter (Fig. 2). Addition of the native BAC element (Z-BAC) to the −50 CaMV 35S promoter enhanced expression 13-fold in ethylene-treated AZ. The orientation of the Z-BAC elements was critical to achieving this high level of expression (Fig. 2). Moreover, expression in AZ was 3-fold higher than the expression level in stems (Fig. 2). In addition, expression was inhibited approximately 40% by auxin (Fig. 2B). A 40% inhibition by auxin is similar to the level of inhibition measured for the wild-type 2.9-kb BAC promoter (Fig. 1B). This suggests that the 18-bp Z-BAC element includes sequence for at least part of the auxin regulation of the BAC promoter. Interestingly, when auxin was applied to AZ and stems bombarded with the wild-type Z-BAC elements in reverse orientation (5-ZIP-R), expression was higher than when the same reverse orientation was used in explants treated with ethylene alone (Fig. 2B). This supports a role for the Z-BAC element where its orientation relative to the start of transcription is critical to its hormonal and abscission-specific regulation in the BAC promoter.
Also of interest here is that expression of the internal control, e35S::GUS, was enhanced at least 3-fold in AZ compared with stems. This most likely reflects the smaller size of cells and higher metabolic activity of the AZ cells relative to stem cells (Sexton and Roberts, 1982). Normalizing the expression of our test BAC constructs to the expression of the e35S::GUS internal control greatly reduces the variability between replicates and it also reduces the difference in expression levels between AZ and stems. For example, the 3-fold enhanced expression of the −50 35S, Z-BAC construct (5-ZIP-F) in AZ compared with stems (Fig. 2) would be approximately 9-fold if not normalized to the internal control. A similar argument applies to the NBD and auxin treatments wherein the e35S::GUS expressions were reduced by 23% and 32%, respectively (Fig. 3B). The NBD and auxin reduction in e35S:GUS expression is most likely because of their inhibition of abscission, which is a very metabolically active process (Sexton and Roberts, 1982). For example, normalization of the data for auxin treated tissues showed that most of the mutated BAC elements were inhibited by an average of 50%; however, if the absolute values were graphed, the auxin inhibition would be closer to 70% inhibition (Fig. 1B). Nevertheless, normalization of data is important not simply for reduction of variability but also because it removes expression enhancements that are not directly related to the cis-acting element being tested, e.g. cell size, metabolic activity, etc.
Of further interest is that the −50 and −90 35S minimal promoters without addition of BAC sequence each displayed approximately 2-fold higher expression in AZs than stems (Fig. 2). It is worth emphasizing that like the other constructs, the expression from the minimal CaMV 35S constructs were normalized to the expression of the internal control, e35S::GUS, which was itself enhanced 3-fold in AZ relative to stems. Therefore, the absolute expression levels for these minimal constructs are approximately 6-fold higher in AZ than stems. The unexpected 2-fold enhancement of the minimal 35S promoters in AZ compared with stems may have important implications for understanding proximal cis-acting elements within the CaMV 35S promoter itself.
The bZIP family of transcription factors binds with differing affinities to a number of related sequences that include the ACGT core motif (Izawa et al., 1993). Nucleotides on either side of the core motif affect the affinity for binding of the different classes of bZIP proteins (Izawa et al., 1993). The Z-BAC element includes an ACGTCA motif that was originally described in the wheat (Triticum aestivum) hex-1 motif, which influences transcription of wheat histone genes (Mikami et al., 1987). The reverse complement of a closely related motif (TGACG), referred to as a TGA-type bZIP-binding site, is repeated twice in the as-1 (activation sequence-1) element identified in the CaMV 35S promoter between −58 and −90 nucleotides from the start of transcription (Lam et al., 1989; Schindler et al., 1992). Transient expression reported here for the −90 constructs was revealing in terms of the enhanced abscission-specific expression supported by the Z-BAC sequence compared with that for the CaMV 35S as-1 element in the −90 35S construct (Fig. 2, B and C). Moreover, the observation that auxin inhibited expression from the Z-BAC minimal construct, 5-ZIP-F, but slightly enhanced expression when the Z-BAC element was in reverse orientation, 5-ZIP-R, is also revealing in terms of how auxin regulates expression from this element (Fig. 2B). The reverse orientation of the Z-BAC element is the same orientation as the TGA motifs in the as-1 element. In this context, it is interesting that auxin has been demonstrated to enhance as-1-dependent transcription (Pascuzzi et al., 1998).
A major component of the ASF-1-binding factor that binds to the as-1 element (Lam et al., 1989) is a TGA-type class II bZIP protein (Niggeweg et al., 2000a). Higher affinity binding to the Z-BAC+ of a class I TGA-type bZIP protein, PvTGA1.1, compared with two class II proteins, PvTGA2.1 and PvTGA2.2 (Fig. 4), is in contrast to that for the as-1 element that favors binding of a class II protein (Niggeweg et al., 2000a). High-affinity binding of class I compared with class II proteins may be an important difference between the Z-BAC and as-1 elements that is also manifested in a difference in the AZ-specific and auxin-regulated expression of the Z-BAC element (Fig. 2).
The involvement of TGA-type bZIP transcription factors in plant development is well documented (Izawa et al., 1993) and, although there have been descriptions of bZIP transcription factors involved in auxin-inducible (Pascuzzi et al., 1998), pathogen-induced (PR), and salicylic acid-regulated gene expression (Despres et al., 2000; Zhou et al., 2000), we are unaware of any reports of bZIP factors acting independently to direct ethylene-induced gene expression. However, the Arabidopsis ocs-binding factor (OBF4), a bZIP protein, and an ethylene-responsive element-binding protein (AtEBP) form a complex that activates the ethylene-inducible PR promoter from the tobacco PRB-1b gene (Buttner and Singh, 1997). Also of interest in this regard is the NPR1 gene, which was originally identified as an Arabidopsis mutant that was compromised in its ability to evoke a systemic acquired resistance response (Despres et al., 2000). The NPR1 protein interacts with a class II TGA-type bZIP factor to increase the binding affinity of the bZIP factor to the as-1 element and the LS5 and LS7 elements in the PR-1 gene promoter (Zhang et al., 1999; Despres et al., 2000). It seems likely, based on precedent and the results reported here for the expression patterns for the PvTGA genes in AZ (Fig. 5), that a TGA-type bZIP factor must complex with or be modified by some other factor that provides the hormonal and tissue specificity expected for regulation at the Z-BAC element.
Similarities between the regulation of BAC and PR gene expression are probably more than simple coincidence. PR gene expression is commonly evoked during abscission as a potential defense against opportunistic infection of vulnerable cells (del Campillo and Lewis, 1992). Therefore, it might be expected that regulation of PR and BAC gene expression may share features in common because induction of these genes during abscission is concomitantly regulated. Nevertheless, transcriptional regulation of PR genes is not expected to be identical to that for the BAC gene because PR gene expression is not generally limited to the AZ nor does it have the same hormonal regulation when expressed in non-AZ tissues (del Campillo and Lewis, 1992; Patterson, 2001).
CONCLUSION
The data presented here provide evidence for the existence of cis-elements in the proximal BAC promoter responsible for both positive and negative regulation of BAC gene expression (Figs. 1 and 2). The 18-bp Z-BAC element that we focused on in the current study is more than a simple enhancer. The Z-BAC element contains sequence information for hormonal and abscission-specific expression (Fig. 2). In addition, we identified three TGA-type bZIP transcription factors that are potential candidates for involvement in the regulation of abscission-specific gene expression (Table I; Fig. 4). Evidence in support of multiple proteins recognizing the same BAC promoter sequence comes from DNA-binding experiments with crude nuclear extracts from ethylene-induced and uninduced AZ where the EMSA displayed a complex banding pattern indicating that more than one factor recognizes the BAC element (data not shown). Additional research is necessary to further characterize the negative and positive cis-elements in the proximal BAC promoter and the trans-acting factors that bind to them. Studies are also needed to identify regulatory proteins that may interact with the TGA-type bZIP transcription factors that bind to the Z-BAC element.
MATERIALS AND METHODS
Plant Material
Bean (Phaseolus vulgaris) cv Red Kidney seeds (Wetsel Seed, Harrisonburg, VA) were germinated in perlite for 12 to 15 d in the greenhouse until the primary leaves were fully expanded and the secondary pinnate leaf just beginning to open. At this time, the leaf blades for the primary leaves were removed and plants harvested by cutting 1 cm above the soil. Harvested plants were surface sterilized in 10% (w/v) commercial bleach for 75 s.
Preparation of Gene Constructs Used for Particle Gun Bombardment
The firefly luciferase open reading frame (ORF) in the pDO432 plasmid (Ow et al., 1986) was used to create the progenitor BAC promoter construct BKm3 (Koehler et al., 1996; Fig. 1A). A luciferase reporter gene was used to maximize the assay sensitivity (Ow et al., 1986). The BKm3 construct was created as a translational fusion of a 2.9-kb BAC promoter, 5′-untranslated region, and ATG start of translation fused in frame directly after the start for translation of the luciferase gene. In addition, the NOS termination sequence in pDO432 was replaced with the BAC termination sequence (Koehler et al., 1996; Fig. 1A).
Site-directed mutagenesis was performed using the Chameleon Double-Stranded Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA). A series of 15 10-bp poly(A+) substitutions was created in the BAC promoter at intervals of 10 bp, beginning at the TATA-box at −23 and ending at position −172. Oligos used for the substitutions consisted of 10 adenine nucleotides flanked by 10 nucleotides of sequence complementary to native BAC sequence on either side of the 10 bp being replaced. The mut ZIP construct was created in a similar fashion, but with only two nucleotide substitutions. All mutations were confirmed by primer-initiated sequencing.
Two additional constructs were prepared for use as internal controls in the particle bombardment experiments. RKm3 was prepared by replacing the luciferase ORF in BKm3 with the GUS ORF from pBI221 (Jefferson et al., 1987). In the other construct, an enhanced CaMV 35S promoter (Kay et al., 1987) was used to replace the 35S promoter in pBI221 to create the constitutively expressed GUS reporter construct e35S::GUS (Fig. 3A).
To create the minimal −90 35S construct, the 5′-upstream sequence starting at nucleotide position −90 of the CaMV 35S promoter in pDO432 was deleted by endonuclease digestion with HindIII and EcoRV and replaced with a multiple cloning site (Fig. 2A). A combination of endonuclease digestion and PCR were used to delete the sequence between −50 and −90 of the −90 35S construct to generate the −50 35S construct. Oligonucleotides consisting of three tandem repeats of native (−113 to −96, TTACTGCACGTCACTCTA) and mutant bZIP-BAC sequence flanked by EcoRI and XbaI restriction sites were synthesized and inserted in the multiple cloning site in the −50 and −90 35S constructs to generate the constructs displayed in Figure 2A.
Transient Expression Assay
Preparation of explants and transient expression assays were similar to those previously described (Koehler et al., 1996) with only a few adaptations. In brief, AZ explants were prepared by cutting the stem 1.0 cm below the first leaf node and then again at the leaf node through the base of the petiole and across the stem to expose a surface that bisects the AZ on either side of the stem. Petiole and stem explants were prepared by excising 1.0-cm sections from the remaining stem and petiole tissue. Explants were positioned upright with the most distal (relative to the parent plant) cut surface facing up in 1.0% (w/v) agar petri plates containing 50 μg mL−1 ampicillin. Each plate contained nine AZs, five stems and 10 petioles, or nine stem explants arranged centrally on the plate. Each particle bombardment session included either three replica plates that were shot only once (Fig. 1B) or two replica plates that were shot twice to increase the number of transformed cells per plate (Figs. 2B and 3C). Each bombardment session, which included two or three replica plates, was repeated at least twice. The controls in Figure 1B reflect the average of more than 15 separate bombardment sessions. Moreover, to further reduce variability, all the test constructs (BAC mutants and minimal Z-BAC constructs) utilized a luciferase reporter gene to take advantage of the high sensitivity of the light-emitting luciferase assay and the very low background emissions from control (wild-type) plant extracts (Ow et al., 1986).
In addition to the precautions noted above, an internal control was also used to normalize the data. The particle load penetrating the tissue varies from one bombardment session to the next. Inclusion of an internal control with each plate bombardment made it possible to normalize the expression level of the test construct to that of the internal control (Koehler et al., 1996). One of the internal controls, RKm3::GUS, is essentially the same as the BKm3 construct (Fig. 1) except that GUS (Jefferson et al., 1987) replaces luciferase as the reporter gene. The RKm3::GUS construct was used only for the ethylene-treated AZ results displayed in Figure 1B to maximize detection of differences in expression levels between the mutated BKm3::luciferase constructs and the wild-type RKm3::GUS construct. For all the other bombardments, including treatments with auxin or NBD and bombardment of stems or petioles, the e35S::GUS construct was used as an internal control (Koehler et al., 1996). Unlike the RKm3::GUS internal control, the e35S::GUS internal control is less responsive to auxin and NBD and highly expressed in stems, petioles, and AZs (Fig. 3B). In addition to these internal controls, each bombardment session included expression data for an external control that consisted of ethylene-treated AZ explants that were bombarded with the wild-type BAC promoter::luciferase construct (BKm3) plus the appropriate GUS internal control. Expression levels for the mutations and the different tissues and treatments are relative to the expression level of this external control (Fig. 1B).
Equal molar concentrations of the BAC::luciferase and RKm3::GUS or e35S::GUS constructs were coprecipitated (0.2 pmol per bombardment) onto 1.6-micron gold microcarriers. To assure that molar ratios were constant, the plasmid DNA was quantified spectrophotometrically at 260 nm and then each of the 17 DNA mixtures was endonuclease digested and visually compared for ethidium bromide staining in an agarose gel. Moreover, many of the DNA samples were also quantified using a fluorometric assay with the Hoechst dye (Bio-Rad, Richmond, CA). Comparison of data from the different assays indicated that the ratio and concentration of the constructs were very close (<10% variability among the samples). A Biolistic PDS-1000/He particle gun (Bio-Rad) was used to bombard explants. Bombardment parameters included a helium pressure of 1,350 psi, one-fourth-inch distance between the rupture disc and macrocarrier, and 5-cm distance between the stopping screen and the plate.
Where indicated, auxin was applied to explants by pipetting 2.0 μL of a 50 mg mL−1 mannitol solution containing 100 μm 2,4-D onto the cut surface of the explants immediately after bombardment. Petri plates containing explants were placed inside 9-L desiccator jars and sealed. Ethylene was injected through a vaccine cap into all the jars to obtain a final concentration of 1.0 μL L−1. NBD was injected into the appropriate jars to obtain a final concentration of 5,000 μL L−1. Jars were held at 25°C in the dark. After 24 h, the jars were opened, allowed to aerate for several seconds, closed, and sealed, and the appropriate concentrations of ethylene and NBD reapplied for another 24 h. Forty-eight hours after bombardment, thin sections (approximately 1 mm) were sliced from the bombarded surfaces of the explants and frozen together in liquid nitrogen. Protein was extracted from frozen tissue by homogenization (3 mL g−1 tissue) in 0.1 m sodium phosphate (pH 7.8), 0.2% (w/v) Triton X-100, 2 mm EDTA, 1 mm dithiothreitol, and 0.1% (w/v) bovine serum albumin. Fluorometric assays for GUS activity were performed as described by Jefferson et al. (1987), except that samples were heated to 55°C for 30 min to inhibit endogenous GUS-like activities. Luciferase was assayed as described in the Luciferase Assay Guide (Analytical Luminescence Laboratory, San Diego) using a luminometer (Bertold, Pittsburgh).
cDNA Library Preparation
Total RNA was isolated from bean leaf AZ that had been exposed to 25 μL L−1 ethylene for 0 and 48 h, i.e. RNA was extracted from both induced and uninduced AZs. The RNA samples were combined and poly(A+) RNA isolated from total RNA using the Oligotex mRNA isolation kit (Qiagen USA, Valencia, CA). A cDNA library in HybriZAP2.1XR (Stratagene) was prepared according to the manufacturer's recommendations using 5 μg of poly(A+) RNA. The titer of the primary library was 1.2 × 107 plaque-forming units (pfu) mL−1. The primary library was amplified to give a final titer of 2.5 × 1010 pfu mL−1.
RT-PCR with Degenerate Oligonucleotides
The peptide sequences of several plant TGA-type bZIP proteins were aligned and a set of nested degenerate oligonucleotides was designed to anneal within the highly conserved basic domain. First strand cDNAs were generated with oligo(dT) using total RNA extracted from ethylene-treated bean leaf AZs. Primers used for subsequent PCR amplification were oligo(dT), TGADG1 (GDYTBGCHCARAAYCGHGAGGC), or TGADG2 (GDYTBGCHCARAAYCGHGAAGC). A second round of PCR used the nested primers TGADG3 (AAAAGYMGHTTRMGVAARAAAGC) and TGADG4 (AAAAGYMGHTTRMGVAARAAGGC). Specific PCR products were eluted, cloned into pGEM-T (Promega, Madison, WI), and sequenced. Subsequently, full-length cDNA clones of PvTGA1.1, PvTGA2.1, and PvTGA2.2 were isolated from the bean AZ cDNA library by conventional screening methods.
EMSAs
The complete coding sequences of PvTGA1.1, PvTGA2.1, and PvTGA2.2 were transferred to pGADT7 (CLONTECH Laboratories, Palo Alto, CA) for in vitro transcription and translation from the T7 RNA polymerase promoter. Proteins were synthesized using the TNT quick-coupled transcription/translation system (Promega). The protein products were labeled with 35S-Met and resolved by SDS-PAGE to ensure efficient synthesis of the factors. EMSAs were performed using double-stranded oligonucleotides of the BAC promoter (−116 to −94) Z-BAC+ (TCATTACTGCACGTCACTCTATCT) and Z-BACm+ (TCATTACTGCAaaTCACTCTATCT). The sense strand oligonucleotide (100 ng) was end labeled with γ32P-[ATP] using T4 polynucleotide kinase (New England Biolabs, Beverly, MA). After removal of unincorporated nucleotides using a G-25 spin column (Amersham, Buckinghamshire, UK), 500 ng of antisense oligonucleotide was added. The oligonucleotide mixture was heated to 70°C for 10 min, then allowed to cool slowly to room temperature. The specific activity ranged from 0.9 to 2.0 × 105 cpm ng−1. EMSA reactions were performed at 30°C for 20 min in a 10-μL solution containing 20 mm HEPES-KOH (pH 7.9), 100 mm KCl, 10 mm MgCl2, 2 mm dithiothreitol, 16% (v/v) glycerol, 0.5 μg of poly dI-dc, 1 μL (70,000 cpm) of labeled oligonucleotide, and 1 μL of in vitro-synthesized protein. Competition experiments included the addition of 100-fold excess unlabeled oligonucleotide and a 10-min incubation at 30°C before addition of the labeled probe. The EMSA reactions were loaded onto a non-denaturing, 4% (w/v) polyacrylamide gel (80:1 [w/v] acrylamide:bisacrylamide in 0.5× Tris-borate/EDTA buffer) and run at 100 V for 45 min. After electrophoresis, gels were blotted onto 3MM filter paper (Whatman, Clifton, NJ), dried under vacuum at 80°C for 45 min, and then exposed to BioMax-MS film (Eastman-Kodak, Rochester, NY) at −70°C.
RNA Gel-Blot Analysis
LAZs and stem explants were treated as previously described (Koehler et al., 1996) with 25 μL L−1 ethylene alone or combinations with 10−5 m IAA in a lanolin paste 4 h before exposure to ethylene and/or 5,000 μL L−1 NBD. Total RNA was isolated from bean LAZ, stem, root, or leaf tissue using a hexadecyltrimethyl ammonium bromide (Sigma, St. Louis) method as described by Hamilton et al. (1995). For RNA-blot analysis, 20 μg of total RNA was fractionated on a gel containing 1% (w/v) agarose, 3% (v/v) formaldehyde, and 20 mm sodium phosphate buffer (pH 6.5). The RNA was transferred to a Hybond N+ nylon membrane (Amersham) and hybridized to 32P-labeled DNA probes of the excised cDNA. Hybridization was carried out at 50°C for 18 h in a solution containing 50% (v/v) formamide (Sigma), 5× Denhardt's reagent, 5× sodium chloride/sodium phosphate/EDTA buffer, 1% (w/v) SDS, and 100 μg mL−1 denatured salmon sperm DNA. The final wash of membranes was at 55°C in 0.1× sodium chloride/sodium phosphate/EDTA buffer and 0.1% (w/v) SDS for 60 min. Washed membranes were exposed to BioMax-MS film (Eastman-Kodak) at −70°C.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
ACKNOWLEDGMENTS
The authors would like to acknowledge Dr. Jonathan Arias (Center for Applied Biotechnology, University of Maryland, College Park) for assistance with the EMSAs, and Vanessa Thai (U.S. Department of Agriculture/Agricultural Research Service/Soybean Genomics and Improvement Lab, Beltsville, MD) for technical assistance.
Footnotes
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.007971.
LITERATURE CITED
- Buttner M, Singh KB. Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC-box DNA-binding protein interacts with an ocs element binding protein. Proc Natl Acad Sci USA. 1997;94:5961–5966. doi: 10.1073/pnas.94.11.5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Campillo E, Lewis LN. Identification and kinetics of accumulation of proteins induced by ethylene in bean abscission zones. Plant Physiol. 1992;98:955–996. doi: 10.1104/pp.98.3.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despres C, DeLong C, Glaze S, Liu E, Fobert PR. The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell. 2000;12:279–290. [PMC free article] [PubMed] [Google Scholar]
- Hamilton AJ, Fray RG, Grierson D. Sense and antisense inactivation of fruit ripening genes in tomato. Cur Topics Microbiol Immunol. 1995;197:77–89. doi: 10.1007/978-3-642-79145-1_6. [DOI] [PubMed] [Google Scholar]
- Izawa T, Foster R, Chua N-H. Plant bZIP protein DNA binding specificity. J Mol Biol. 1993;230:1131–1144. doi: 10.1006/jmbi.1993.1230. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusion: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri F, Lam E, Chua N-H. Two tobacco DNA-binding proteins with homology to the nuclear factor CREB. Nature. 1989;340:727–730. doi: 10.1038/340727a0. [DOI] [PubMed] [Google Scholar]
- Kay R, Chan A, Daly M, McPherson J. Duplication of CaMV 35S promoter sequences creates a strong enhancer for plant genes. Science. 1987;236:1299–1302. doi: 10.1126/science.236.4806.1299. [DOI] [PubMed] [Google Scholar]
- Koehler DE, Lewis LN, Shannon LM, Durbin ML. Purification of abscission zone cellulase. Phytochemistry. 1980;20:409–412. [Google Scholar]
- Koehler SM, Matters GM, Nath P, Kemmerer EC, Tucker ML. The gene promoter for a bean abscission cellulase is ethylene-induced in transgenic tomato and shows high sequence conservation with a soybean abscission cellulase. Plant Mol Biol. 1996;31:595–606. doi: 10.1007/BF00042232. [DOI] [PubMed] [Google Scholar]
- Lam E, Benfey PN, Gilmartin PM, Fang RX, Chua N-H. Site-specific mutations alter in vitro factor binding and change promoter expression pattern in transgenic plants. Proc Natl Acad Sci USA. 1989;86:7890–7897. doi: 10.1073/pnas.86.20.7890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis LN, Varner JE. Synthesis of cellulase during abscission of Phaseolus vulgaris leaf explants. Plant Physiol. 1970;46:194–199. doi: 10.1104/pp.46.2.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami K, Nakayama T, Kawata T, Tabata T, Iwabuchi M. Specific interaction of nuclear protein HBP-1 with the conserved hexameric sequence ACGTCA in the regulatory region of wheat histone genes. Plant Cell Physiol. 1987;30:107–119. [Google Scholar]
- Niggeweg R, Gatz C. Isolation of TGA2.1, a member of a new subclass of the TGA family of bZIP transcription factors in Nicotiana tabacum. Plant Physiol. 1997;113:1464. [Google Scholar]
- Niggeweg R, Thurow C, Kegler C, Gatz C. Tobacco transcription factor TGA2.2 is the main component of as-1-binding factor ASF-1 and is involved in salicylic acid- and auxin-inducible expression of as-1-containing target promoters. J Biol Chem. 2000a;275:19897–19905. doi: 10.1074/jbc.M909267199. [DOI] [PubMed] [Google Scholar]
- Niggeweg R, Thurow C, Weigel R, Pfitzner U, Gatz C. Tobacco TGA factors differ with respect to interaction with NPR1, activation potential and DNA-binding properties. Plant Mol Biol. 2000b;42:775–788. doi: 10.1023/a:1006319113205. [DOI] [PubMed] [Google Scholar]
- Ow DW, Wood KV, DeLuca M, DeWet JR, Helinski DR, Howell SH. Transient and stable expression of the firefly luciferase gene in plant cells and transgenic plants. Science. 1986;234:856–859. doi: 10.1126/science.234.4778.856. [DOI] [PubMed] [Google Scholar]
- Pascuzzi P, Hamilton D, Bodily K, Arias J. Auxin-induced stress potentiates trans-activation by a conserved plant basic/leucine-zipper factor. J Biol Chem. 1998;273:26631–26637. doi: 10.1074/jbc.273.41.26631. [DOI] [PubMed] [Google Scholar]
- Patterson SE. Cutting loose: abscission and dehiscence in Arabidopsis. Plant Physiol. 2001;126:494–500. doi: 10.1104/pp.126.2.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler U, Beckmann H, Cashmore AR. TGA1 and G-box binding factors: two distinct classes of Arabidopsis leucine zipper proteins compete for the G-box-like element TGACGTGG. Plant Cell. 1992;4:1309–1319. doi: 10.1105/tpc.4.10.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton R, Roberts JA. Cell biology of abscission. Annu Rev Plant Physiol. 1982;33:133–162. [Google Scholar]
- Sisler EC, Goren M, Huberman M. Effect of 2,5-norbornadiene on abscission and ethylene production in citrus leaf explants. Physiol Plant. 1985;63:114–120. [Google Scholar]
- Tucker ML, Baird SL, Sexton R. Bean leaf abscission: tissue-specific accumulation of a cellulase mRNA. Planta. 1991;186:52–57. doi: 10.1007/BF00201497. [DOI] [PubMed] [Google Scholar]
- Tucker ML, Sexton R, del Campillo E, Lewis LN. Bean abscission cellulase: characterization of a cDNA clone and regulation of gene expression by ethylene and auxin. Plant Physiol. 1988;88:1257–1262. doi: 10.1104/pp.88.4.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Foley RC, Singh KB. Isolation and characterization of two related Arabidopsis ocs-element bZIP binding proteins. Plant J. 1993;4:711–716. doi: 10.1046/j.1365-313x.1993.04040711.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Fan W, Kinkema M, Li X, Dong X. Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of PR-1 gene. Proc Natl Acad Sci USA. 1999;96:6523–6528. doi: 10.1073/pnas.96.11.6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JM, Trifa Y, Silva H, Pontier D, Lam E, Shah J, Klessig DF. NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol Plant-Microbe Interact. 2000;13:191–202. doi: 10.1094/MPMI.2000.13.2.191. [DOI] [PubMed] [Google Scholar]