Abstract
Assembly of two orthologous proteins associated with meiotic chromosome axes in Arabidopsis thaliana (Asy1 and Zyp1) was studied immunologically at meiotic prophase of meiosis of wild-type rye (Secale cereale) and its synaptic mutant sy10, using antibodies derived from A. thaliana. The temporal and spatial expression of the two proteins were similar in wild-type rye, but with one notable difference. Unlike A. thaliana, in which foci of the transverse filament protein Zyp1 appear to linearize commensurately with synapsis, linear tracts of Asy1 and Zyp1 protein form independently at leptotene and early zygotene of rye and coalign into triple structures resembling synaptonemal complexes (SCs) only at later stages of synapsis. The sy10 mutant used in this study also forms spatially separate linear tracts of Asy1 and Zyp1 proteins at leptotene and early zygotene, and these coalign but do not form regular triple structures at midprophase. Electron microscopy of spread axial elements reveals extensive asynapsis with some exchanges of pairing partners. Indiscriminate SCs support nonhomologous chiasma formation at metaphase I, as revealed by multi-color fluorescence in situ hybridization enabling reliable identification of all the chromosomes of the complement. Scrutiny of chiasmate associations of chromosomes at this stage revealed some specificity in the associations of homologous and nonhomologous chromosomes. Inferences about the nature of synapsis in this mutant were drawn from such observations.
THE availability of advanced genomic and proteomic resources in tractable model organisms, such as yeast and Arabidopsis thaliana, has provided unprecedented access to the genes and proteins involved in the control of meiosis. This functional genomic infrastructure has also precipitated detailed comparisons of meiosis in closely and distantly related organisms, not only with the intention of isolating orthologs with key roles in the process, but also with the goal of assaying the degree of similarity of structure and function of key meiotic genes and proteins of organisms across the phylogenetic spectrum. Such comparisons have revealed that meiosis is conserved, insofar as a number of meiotic genes appear to have orthologs in a range of different organisms (for reviews see Zickler and Kleckner 1998, 1999; Bogdanov 2003; Bishop and Zickler 2004; Page and Hawley 2004; Gerton and Hawley 2005). While comparisons may point to common pathways of control of meiotic processes, structural similarity between orthologous genes and the proteins that they encode does not necessarily indicate equivalence of function. For example, the HOP1 gene of budding yeast encodes a protein that is needed for axial element formation during synapsis (Hollingsworth and Byers 1989; Zickler and Kleckner 1999), whereas its ortholog in A. thaliana (ASY1) encodes a protein associated with chromatin in close proximity to the synaptonemal complex (SC) (Caryl et al. 2000). Conversely, some proteins appear to perform the same functions in different organisms, yet have little amino acid sequence in common. For example, the transverse filament protein of the SC of A. thaliana (Zyp1; Higgins et al. 2005) shares only 18–20% sequence identity and 36–40% similarity with the corresponding protein Zip1 from Saccharomyces cerevisiae (Sym et al. 1993), with Scp1 from mammals (Heyting 1996), and with C(3)G from Drosophila melanogaster (Page and Hawley 2004). Furthermore, there are other proteins that appear to be unique to particular organisms, such as the Phs1 protein of maize, which has no significant homology to known proteins, apart from superficial resemblance to two families of helicases from fungi (Pawlowski et al. 2004). Taken as a whole, these observations emphasize the fact that an understanding of meiosis requires an integration of our knowledge of the process in different organisms (Shaw and Moore 1998).
Rye (Secale cereale) is a close relative of wheat and therefore an excellent cytological model for investigating meiosis and recombination in the temperate cereals and grasses (Jenkins et al. 2005). It has a nuclear genome, which is replete with gene duplicates and reiterated, noncoding DNA (Devos et al. 1993; Devos and Gale 2000; Alkhimova et al. 2004; Schulman et al. 2004; Varshney et al. 2004) and which is 64 times the size of the compact genome of A. thaliana. To assay the extent of similarity of meiosis in these two divergent plant species, and to relate any differences to vastly different genome architectures, it was decided to focus upon the expression and recruitment of meiosis-specific proteins associated with the SC, a structure that may be considered a paradigm for the assembly and interaction of structural and recombinogenic proteins. More specifically, this article describes primarily the immunolocalization of two SC-related proteins (Asy1 and Zyp1) at meiotic prophase of wild-type rye and a synaptic mutant sy10, using antibodies derived from A. thaliana. The mutant was chosen from a collection of spontaneous mutants (Sosnikhina et al. 2005) on the basis of its largely asynaptic phenotype and propensity to synapse chromosomes indiscriminately (Fedotova et al. 1994; Sosnikhina et al. 1994b). Although this mutant has a very low chiasma frequency, it forms some bivalents and characteristic “sticky” associations at first metaphase. Simultaneous fluorescence in situ hybridization (FISH) with five landmark DNA probes enabled all 14 chromosomes to be identified with a high degree of confidence. This permitted for the first time an analysis of chiasmate and nonchiasmate associations at this stage, from which inferences could be drawn about the relationship between chromosome structure and pairing behavior when homology, recognition, and synapsis are compromised by mutation. Taken together, the cytological and molecular data contribute to the assembly of a phenotypic “identikit” for this mutant and aid the reconstruction of genetic control pathways of meiosis in rye. The study also validates the utility of A. thaliana as a model for meiosis across the divide between the monocots and the dicots.
MATERIALS AND METHODS
Plant material:
Spontaneous synaptic mutation sy10 of winter rye (S. cereale L. 2n = 2x = 14) was originally isolated in 1985 from selfed F1 hybrids of crosses between individual plants of Trans-Caucasian weedy rye and plants of self-fertile inbred lines (Sosnikhina et al. 1994a). Homozygotes for sy10 are largely sterile and are isolated from segregating progenies of selfed heterozygotes on the basis of their aberrant meiotic phenotype, high univalency at metaphase I, and distinctive indiscriminate synapsis during meiotic prophase (Fedotova et al. 1994; Sosnikhina et al. 1994b). Segregating progenies studied in Aberystwyth originate from a single plant grown and selfed in 1999 in an experimental field at St. Petersburg, representing the third generation of inbreeding following an intermediate cross of one plant with Secale kuprijanovii Grossh. The progeny of this single plant were sown in an unheated greenhouse in Aberystwyth in October 2002 and allowed to flower in season (May to June 2003). All subsequent progeny displayed a more extreme form of the sy10 phenotype (analogous to “group II”), which has a higher mean univalent frequency at metaphase I compared with “group I” (Fedotova et al. 1994; Sosnikhina et al. 1994b). Segregating progeny used for the FISH study originated from a single sib progenitor of the Aberystwyth stocks. To gain access to meiotic material for other experiments the year round, and to standardize growth and flowering conditions, small batches of seed of segregating sy10 families have been sown regularly since 2004, and 3-day-old seedlings were vernalized artificially for 40 days in a dark refrigerator at 0°. Following the cold treatment, the seedlings were transferred to pots in a transgenic greenhouse and grown to maturity under 16-hr days with 60 μmol/m2/sec illumination at 15°, and 10° nights. Individuals with high bivalent frequencies segregating in the sy10 families were used as controls. The spring rye strain Shkolnaya Hl [Ddw1–Dominant dwarfness 1; Leningrad region, K-11575, N. I.Vavilov All-Russian Research Institute of Plant Industry (VIR), St. Petersburg, Russia], which has no vernalization requirement, was also grown regularly under the same greenhouse conditions to provide an additional and ready source of control meiotic material. Spikelets from meiotic inflorescences either were fixed individually in a fresh 3:1 (v/v) mixture of ethanol and acetic acid and stored at −20° or were used fresh for surface spreading and immunocytology.
FISH:
FISH of chromosomes at metaphase I of meiosis was performed essentially as described by Mikhailova et al. (2001), but with the following modifications. Anthers were digested in an enzyme mixture comprising 0.3% (w/v) cellulase (Onozuka RS), 0.3% (w/v) pectolyase Y-23 (Seishin Pharmaceutical), and 0.3% cytohelicase (Sigma, St. Louis) in 10 mm citrate buffer (pH 4.5). pSc200 is a 521-bp insert in pUC18 comprising a 380-bp tandem repeat unit of subtelomeric DNA from rye (Vershinin et al. 1995). The insert was amplified and labeled by PCR as described in Mikhailova et al. (2001). The sequence has 13 major subtelomeric sites and 10 minor sites in the haploid complement of this inbred line. pSc250 is a 476-bp insert in pUC18 and is a representative of a family of tandemly organized subtelomeric DNA sequences of rye with an unusually extended monomer length of ∼500 bp (Vershinin et al. 1995). The sequence localizes to 13–14 major subtelomeric sites and six minor sites and is proximal to pSc200. The insert was amplified and labeled by PCR using M13 forward and reverse primers and FluoroLink Cy5-dUTP (Amersham Pharmacia) under the same conditions as for labeling the pSc200 probe. CCS1 is a 260-bp motif (Aragon-Alcaide et al. 1996) of a centromere-specific clone (Hi-10) originally isolated from Brachypodium sylvaticum (Abbo et al. 1995). It is localized exclusively to the pericentromeric regions of all rye chromosomes. It accurately marks centromeres and delimits chromosome arms. The sequence was amplified by PCR in the presence of digoxigenin-11-dUTP (Roche) according to Aragon-Alcaide et al. (1996). 25S rDNA is a 2.3-kb subclone of the 25S rDNA coding region of A. thaliana (Unfried and Gruendler 1990). It was labeled with FluoroRed (rhodamine-4-dUTP) or digoxigenin-11-dUTP (Roche) in two separate reactions by nick translation according to the manufacturer's instructions (Roche). A composite probe was made by mixing equal proportions of the two labeled products. 25S rDNA has a single locus and is a diagnostic feature of the pair of 1R chromosomes. 5S rDNA was derived from the wheat clone pTa 794 (Gerlach and Dyer 1980) and was amplified by PCR in the presence of FluoroRed (rhodamine-4-dUTP) or digoxigenin-11-dUTP (Roche), using the same conditions as for pSc200, and mixed in equal proportions. 5S rDNA has two or three loci in rye and the potential to identify the pairs of chromosomes 1R, 3R, and 5R (Cuadrado et al. 1995; Cuadrado and Jouve 2002). All five probes were purified by precipitation under ethanol. Each probe was mixed to a final concentration of 2–2.5 ng/μl in a hybridization solution containing inter alia 50% formamide and 2× SSC. The probes were hybridized overnight at 37°, and the slides were washed stringently in 0.1× SSC for 30 min at 50°. Digoxigenin was detected with antidigoxigenin antibodies conjugated to fluorescein, and the chromosomes were counterstained with DAPI (0.8 μg/ml).
FISH of chromosomes at early meiotic prophase using the telomeric probe HT100.3 and the centromeric probe INTR2 was performed according to Hajdera et al. (2003), and Langdon et al. (2000), respectively. Both PCR products were mixed directly to a final concentration of 2–2.5 ng/μl in a hybridization solution containing inter alia 50% formamide and 2× SSC. The probes were hybridized overnight at 37°, and the slides were washed stringently in 20% formamide in 0.1× SSC for 10 min at 42°. Digoxygenin was detected with antidigoxigenin antibodies conjugated to fluorescein, and the chromosomes were counterstained with DAPI (1 μg/ml).
Electron microscopy:
Surface-spread meiotic cells were prepared and imaged essentially as described by Chatterjee and Jenkins (1993), but with the following modification. Images were captured using a Jeol 100CX electron microscope equipped with an analytical scanning image device (ASID) and running Printerface for Windows (K. E. Developments).
Immunocytology:
The following method is based upon procedures by Terasawa et al. (1995), Anderson et al. (1994), and Heyting et al. (1994). Fresh anthers at the desired stage of meiosis were extruded onto a siliconized depression slide containing 5 μl of fixative (2% paraformaldehyde in 50 mm potassium phosphate buffer, pH 7.5). Extruded rods of pollen mother cells (PMCs) were cut into small pieces using a fine, sharp needle. Another 5 μl of fixative was added and mixed with the needle and the depression slide was placed in a humid chamber at room temperature for 6 min. An additional 10 μl of fresh fixative was added to the droplet and left in the humid chamber for another 2 min. The suspension of PMCs was transferred onto a dry clean slide and 0.5 μl of 0.4% Triton in PBS was added to the suspension and left for another 2 min. A coverslip was placed on the suspension and lifted gently with the point of a needle to release protoplasts from the callose walls of the PMCs. Excess fixative was removed by gentle squashing and blotting with filter paper. The slides were frozen at −80° and left for 30 min, and their coverslips were removed using a razor blade and placed into PBS. The slides were then rinsed in PBS for 5 min, in 0.1 m ammonium chloride (in PBS) for 5 min, and finally in PBS for another 5 min. The preparations were blocked using 2 ml of 5% milk in PBS blocking buffer per slide at room temperature for 30 min. The blocking buffer was then poured away and the slides were dipped in PBS for 1 sec. The slides were then incubated with primary antibodies diluted with blocking buffer: anti-Asy1 raised in rabbit or rat diluted 1:300 (Caryl et al. 2000) and anti-Zyp1 raised in rabbit or rat diluted 1:150 (Higgins et al. 2005). The slides were then incubated in a humid chamber for 1 hr at room temperature, placed at 4° overnight, and left at room temperature for 1 hr. The primary antibodies were removed by briefly dipping the slides into PBS. The slides were then incubated with secondary antibodies diluted in blocking buffer; Alexa Fluor 488 donkey anti-rabbit immunoglobulin G (IgG) (Molecular Probes, Eugene, OR) diluted to 1:500 and Alexa Fluor 546 goat anti-rat IgG (Molecular Probes) diluted to 1:250. They were incubated in a humid chamber for 2 hr at room temperature. The slides were finally washed four times in PBS for 15 min each. Ten microliters of DAPI (0.5 μg/ml) in Vectashield was applied to the wet slides. The preparations were then covered with coverslips and left for 2–3 hr at room temperature in the dark to evenly distribute the DAPI in Vectashield beneath the coverslips.
Imaging:
Fluorescent images were captured by a Hamamatsu (Bridgewater, NJ) Orca digital camera coupled to a Zeiss Axioplan fluorescence microscope and assigned false color and manipulated uniformly using Wasabi software. Chromosomes at metaphase I and immunofluorescently stained nuclei were optically sectioned using a Bio-Rad (Hercules, CA) MRC-1024 MPR confocal laser scanning microscope, running LaserSharp2000 5.2 image capturing and processing software (Bio-Rad), and recorded in separate green (fluorescein or Alexa Fluor 488), red (rhodamine or Alexa Fluor 546), blue (Cy5), and merged channels. Each DAPI-stained metaphase plate was also imaged with a Zeiss Axioplan fluorescence microscope, recorded on Fujichrome Provia 400 color film with an MC100 camera, and converted digitally to TIFF using a film scanner. Confocal stacks of the immunofluorescently stained nuclei were deconvoluted using Volocity 2.6.1 (Improvision). Information about chromosome identities and characteristics and the nature of their associations was entered for each separate cell into an Access relational database coupled directly to a complete library of images stored in Adobe Photoshop 5.0.
RESULTS
Chromosome associations at metaphase I:
All 100 PMCs from each of three plants of the spring rye control had seven bivalents and an overall mean chiasma frequency of 13.6 ± 0.55. The efficiency of bivalent formation and high levels of recombination are typical for an outcrossing representative of this species. Chiasma frequencies were also scored from 100 PMCs each of 13 wild-type plants from two segregating families in 2005 and of 11 wild-type plants from three segregating families in 2006. χ2 tests for homogeneity, using an expected frequency of 14 chiasmata/cell derived from spring rye, showed no significant variation in chiasma frequencies between plants within each year and across both years (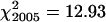 , ν = 12, 0.5 > P > 0.25;
, ν = 12, 0.5 > P > 0.25; 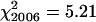 , ν = 10, 0.9 > P > 0.75;
, ν = 10, 0.9 > P > 0.75; 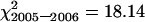 , ν = 23, P > 0.05). This permitted pooling of the data to give a mean bivalent frequency of 6.7 ± 0.23 and a mean chiasma frequency of 10.9 ± 0.89 for wild-type segregants in the sy10 lines. Both these values are significantly lower than those of spring rye and are typical of inbred lines of rye (Rees 1955; Rees and Thompson 1956; Sosnikhina et al. 1994a). This difference necessitated the use of both spring rye and wild-type segregants in sy10 families as controls in this study.
, ν = 23, P > 0.05). This permitted pooling of the data to give a mean bivalent frequency of 6.7 ± 0.23 and a mean chiasma frequency of 10.9 ± 0.89 for wild-type segregants in the sy10 lines. Both these values are significantly lower than those of spring rye and are typical of inbred lines of rye (Rees 1955; Rees and Thompson 1956; Sosnikhina et al. 1994a). This difference necessitated the use of both spring rye and wild-type segregants in sy10 families as controls in this study.
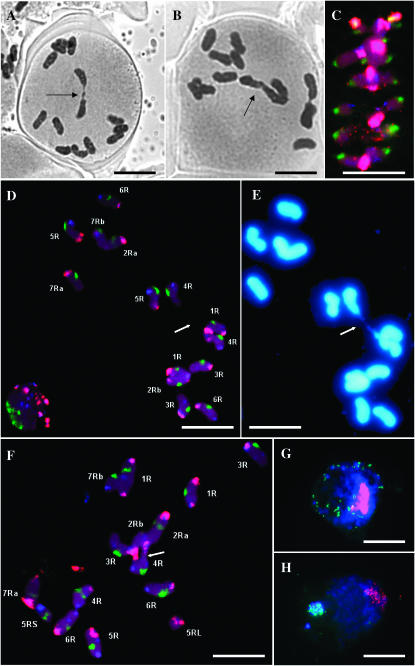
Bivalent and chiasma frequencies were also scored from 2000 to 4000 PMCs each of seven sy10 mutant plants from three segregating families in 2006. The sy10 mutant forms mostly univalents and consequently has a very low bivalent frequency (0.04 ± 0.034) and chiasma frequency (0.04 ± 0.034). χ2 tests for homogeneity, using the maximum of 14 univalents/cell, showed no significant variation in univalent frequency among the seven plants (χ2 = 0.004, ν = 6, P > 0.99). These frequencies are even lower than the more extreme (group II) phenotype of sy10 described by Fedotova et al. (1994) and Sosnikhina et al. (1994b). The predictability and uniformity of the behavior of chromosomes in the mutant validates the use of relatively small subsets of cells from individual plants in the studies described below. Figure 1A shows a typical pollen mother cell at this stage, with one bivalent with a single, distal chiasma and 12 univalents. Univalents are characteristically “sticky” in this mutant and tend to associate in a parallel fashion or end-to-end creating multiple, nonchiasmate complexes (Figure 1B).
Figure 1.—
(A) PMC of sy10 at metaphase I showing a bivalent with a single, distal chiasma (arrow) and 12 univalents. (B) PMC of sy10 at metaphase I showing a “sticky” complex of three univalents (arrow). (C) Projection of an optical stack through a typical PMC at metaphase I of Sy10 wild type. The chromosomes are probed with pSc200 (red), pSc250 (blue), 25S rDNA (yellow), and CCS1 (green). (D) Projection of an optical stack through a PMC at metaphase I of sy10 hybridized in situ with the same probes as in C. All chromosomes are identified. The two chromosomes 4R are connected by a chromatin bridge (arrow) and constitute the only rod bivalent in this cell. (E) PMC in D stained with DAPI, which reveals the chromatin bridge between the two 4R chromosomes. (F) Similar cell to that in D, but showing a single ring bivalent (arrow) between two nonhomologous chromosomes. (G) Typical PMC at premeiotic interphase hybridized in situ with centromeric (red) and telomeric (green) probes and counterstained with DAPI. (H) Similar cell to that in G but at leptotene, showing a tight telomeric cluster (bouquet) colocalizing with DAPI-positive telomeric heterochromatin. Bars, 10 μm.
To determine if the chiasmata and “sticky” associations in this mutant are formed between homologous or nonhomologous chromosomes, the seven pairs of chromosomes were marked with five fluorescent probes. Optical stacks and corresponding DAPI images were taken for 14 chromosome complements at metaphase I of wild type (Figure 1C), and 78 of the mutant. The latter were selected on the basis of successful FISH, ease of analysis, and the presence of rare bivalents revealed with DAPI staining. All of the chromosomes from most of the 78 mutant cells could be identified by the patterns of colored probes, which defined the positions of the centromeres and discriminated almost all chromosome arms (Figure 1D). Using these features, together with published information from Tikhonovich and Fadeyeva (1976), Tikhonovich et al. (1987), Mikhailova et al. (1993), and Alkhimova et al. (1999), the 14 chromosomes of the complement were identified and designated 1R–7R in accordance with the standard international nomenclature (Sybenga 1983). Despite the fact that sy10 is an inbred line, chromosomes 2R and 7R showed heteromorphism with respect to the presence or abundance of the pSc200 and pSc250 repeat sequences. The heteromorphism of the pair of chromosomes 7R was also evident in the wild-type control; chromosome variation of this nature has been reported in other lines of rye (Alkhimova et al. 1999). The structural differences between homologs made identification of chromosomes 4R, 6R, and 7R ambiguous in 18% of cases, but chromosomes 1R, 2R, 3R, and 5R could be identified with 99% confidence.
Identification of each chromosome allows an unambiguous analysis of the chromosomes involved in a chiasma. Figure 1D shows a cell with one homologous rod bivalent involving the two 4R chromosomes connected by a thin chromatin bridge revealed by DAPI staining (Figure 1E). The remainder of the chromosomes are univalent. The cell is similar in these respects to that shown in Figure 1A. There are clearly nonchiasmate, parallel associations between chromosomes 1R and 4R and between chromosomes 1R and 2Rb. Similarly, this unambiguous identification shows that some of the chiasmata are formed between two nonhomologous chromosomes. The cell shown in Figure 1F also has only one bivalent, but this comprises two nonhomologous chromosomes (2Rb and 4R) held together by two distal chiasmata. The rest of the chromosomes are univalent and are engaged in a variety of end-to-end and parallel associations.
To evaluate the relative proportions of homologous and nonhomologous associations, we analyzed the 324 associations recorded in the 78 nuclei of sy10. Table 1 shows clearly that only 37 (or 11.4%) are in fact true chiasmata, reflecting the typically low chiasma frequency in this largely asynaptic mutant. Moreover, of these, only 23 (or 62.2%) are between homologous chromosomes. Strikingly, over half (56%) of these involve the short arm of chromosome 4. This demonstrates not only significant asymmetry of chiasma formation in this chromosome (χ2 = 9.3, ν = 1, 0.01 < P < 0.001), but also significant chromosome-specific formation of homologous chiasmata (χ2 = 30.96, ν = 1, P < 0.001). The latter is reinforced by the observation that no homologous chiasmata were formed between the two 5R chromosomes. Of the nonhomologous chiasmata, we found that over half (54%) also involved the short arm of chromosome 4. This clearly indicates that this arm is more often involved in recombination than all the other chromosome arms of the complement. The only other chromosome showing a greater propensity to recombine than others is 5R (5 of the 37 chiasmata), but only nonhomologously (Table 1) and >50% with 4RS.
TABLE 1.
Chiasmate associations of chromosome arms in 78 PMCs of the sy10 mutant
| Arm | 1RS | 1RL | 2RS | 2RL | 3RS | 3RL | 4RS | 4RL | 5RS | 5RL | 6RS | 6RL | 7RS | 7RL |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1RS | ||||||||||||||
| 1RL | 1(0) | |||||||||||||
| 2RS | 1(0) | |||||||||||||
| 2RL | 1(0) | 1(0) | ||||||||||||
| 3RS | ||||||||||||||
| 3RL | 1(0) | 1(0) | 1(0) | |||||||||||
| 4RS | 13(1) | 3(0) | 1(0) | 1(0) | ||||||||||
| 4RL | 1(0) | |||||||||||||
| 5RS | ||||||||||||||
| 5RL | 1(0) | 1(1) | ||||||||||||
| 6RS | ||||||||||||||
| 6RL | 2(0) | 1(1) | ||||||||||||
| 7RS | 2(0) | |||||||||||||
| 7RL | 4(0) |
The numbers in parentheses indicate how many of the associations are ambiguous due to doubts about chromosome identity. S, short arm; L, long arm.
Finally, analysis of all nonchiasmate associations (Table 2) shows clearly that there is no specific pattern, indicating that chromosomes “stick” randomly to one another by an unknown process. This also appears to hold true for 19 associations of particular centromeres with each other (Table 3) and for 33 associations of specific centromere regions with particular chromosome arms (Table 4). Close examination of 34 associations involving three or more chromosomes indicated that no chromosome is significantly more involved in these multiple associations than others.
TABLE 2.
Nonchiasmate associations of chromosome arms in 78 PMCs of the sy10 mutant
| Arm | 1RS | 1RL | 2RS | 2RL | 3RS | 3RL | 4RS | 4RL | 5RS | 5RL | 6RS | 6RL | 7RS | 7RL |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1RS | 0(0) | 1(0) | 4(0) | 4(0) | 5(0) | 3(0) | 4(0) | 4(1) | 0(0) | 5(0) | 2(2) | 4(0) | 4(1) | 5(2) |
| 1RL | 1(0) | 6(0) | 7(0) | 1(0) | 4(0) | 10(1) | 3(0) | 2(0) | 5(0) | 2(0) | 3(0) | 5(0) | 3(1) | |
| 2RS | 1(0) | 1(0) | 3(0) | 6(0) | 0(0) | 2(1) | 4(0) | 6(0) | 2(0) | 3(0) | 2(0) | 3(0) | ||
| 2RL | 1(0) | 1(0) | 1(0) | 7(2) | 1(0) | 5(0) | 6(0) | 2(0) | 4(0) | 1(0) | 3(1) | |||
| 3RS | 3(1) | 3(2) | 1(0) | 4(0) | 1(0) | 4(0) | 3(0) | 5(2) | 2(0) | 2(0) | ||||
| 3RL | 3(0) | 3(1) | 5(0) | 0(0) | 4(0) | 2(1) | 4(2) | 2(0) | 1(0) | |||||
| 4RS | 3(0) | 1(0) | 5(0) | 5(1) | 2(0) | 8(0) | 2(0) | 1(0) | ||||||
| 4RL | 2(1) | 2(0) | 1(0) | 6(1) | 6(1) | 0(0) | 3(0) | |||||||
| 5RS | 0(0) | 1(0) | 2(1) | 6(0) | 1(0) | 2(0) | ||||||||
| 5RL | 1(0) | 5(1) | 5(0) | 9(3) | 2(0) | |||||||||
| 6RS | 1(1) | 3(1) | 6(1) | 2(0) | ||||||||||
| 6RL | 2(0) | 2(0) | 2(2) | |||||||||||
| 7RS | 3(1) | 2(0) | ||||||||||||
| 7RL | 6(2) |
The numbers in parentheses indicate how many of the associations are ambiguous due to doubts about chromosome identity. Italics indicate which chromosome arms also form chiasmate associations (see Table 1). S, short arm; L, long arm.
TABLE 3.
Centromere–centromere associations in 78 PMCs of the sy10 mutant
| Centromere | 1R | 2R | 3R | 4R | 5R | 6R | 7R |
|---|---|---|---|---|---|---|---|
| 1R | 1 | ||||||
| 2R | 1 | ||||||
| 3R | 1 | ||||||
| 4R | 1 | 1 | 1 | ||||
| 5R | 2 | 2 | 2 | ||||
| 6R | 1 | 1 | 1 | 3 | |||
| 7R | 1 |
There are no ambiguities in chromosome identification.
TABLE 4.
Associations of centromeric regions with specific chromosome arms in 78 PMCs of the sy10 mutant
| Chromosome
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1R
|
2R
|
3R
|
4R
|
5R
|
6R
|
7R
|
|||||||||
| Centromere | Arm: | S | L | S | L | S | L | S | L | S | L | S | L | S | L |
| 1R | 1(0) | 1(0) | |||||||||||||
| 2R | 1(0) | 1(0) | 1(0) | 2(0) | 1(0) | 1(0) | |||||||||
| 3R | 1(0) | 1(0) | 1(0) | ||||||||||||
| 4R | 1(0) | 1(0) | 1(0) | 1(0) | |||||||||||
| 5R | 1(0) | 1(0) | 2(0) | 3(1) | 1(0) | ||||||||||
| 6R | 1(0) | 1(1) | 1(0) | 1(0) | |||||||||||
| 7R | 1(0) | 1(0) | 1(0) | 1(0) | 1(0) | ||||||||||
The numbers in parentheses indicate how many of the associations are ambiguous due to doubts about chromosome identity. S, short arm; L, long arm.
Early association and synapsis of chromosomes:
To interpret the failure and promiscuity of chromosome association at metaphase I in this mutant, the early association and synaptic behavior of chromosomes were scrutinized. Figure 1G shows a typical pollen mother cell at premeiotic interphase in sy10. The centromeres form a single amorphous mass in Rabl orientation, and the telomeres are distributed throughout the nucleus. At early leptotene (Figure 1H), the centromeres are grouped and the telomeres form a tight cluster at the nuclear envelope coincident with DAPI-stained telomeric heterochromatin. The passage of cells into a bouquet conformation appears not to be impaired by the sy10 mutation and appears similar to that reported for the nonallelic mutant sy9 (Mikhailova et al. 2001). Subsequent relative movements of centromeres and telomeres were not the subject of this study, but are currently under investigation.
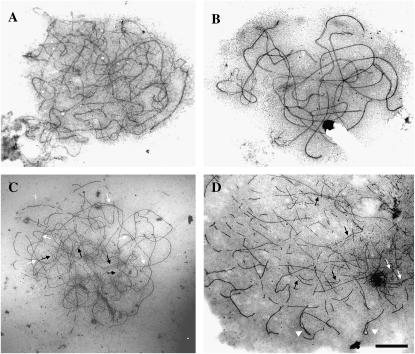
The surface-spread SC complements shown in Figure 2, A and B, represent the normal progression of synapsis from mid- to late zygotene in wild type. The seven virtually complete SCs in Figure 2B indicate that pairing is regular and effective in terms of supporting recombination in this material. By contrast, Figure 2C shows a cell at a comparable stage in the mutant. Its axial elements are largely unsynapsed, with only a few short and discrete segments of alignment to ∼100 nm. Tracing individual axial elements reveals pairing partner switches, indicative of the promiscuous synapsis that typifies this mutant. Synapsis fails to progress beyond this point, as indicated in the asynaptic fragments of axial elements at diplotene (Figure 2D). Clearly, the widespread failure of synapsis and loss of integrity of homologous association are important determinants of the mutant phenotype at metaphase I.
Figure 2.—
Electron micrographs of SCs of Sy10 wild type, showing the normal progression of synapsis from early (A) to late (B) zygotene. (C) Surface-spread PMC of the sy10 mutant at a stage equivalent to zygotene, showing widespread asynapsis and some short stretches of SC (solid arrows). Switches of pairing partners indicating indiscriminate synapsis are delimited by open arrows. (D) Late diplotene in the sy10 mutant showing fragments of axial cores and remnants of SCs (solid and open arrows, respectively) and foldbacks (arrowheads). Bar, 5 μm.
Axis and SC proteins are recruited and associate regularly in the absence of synapsis:
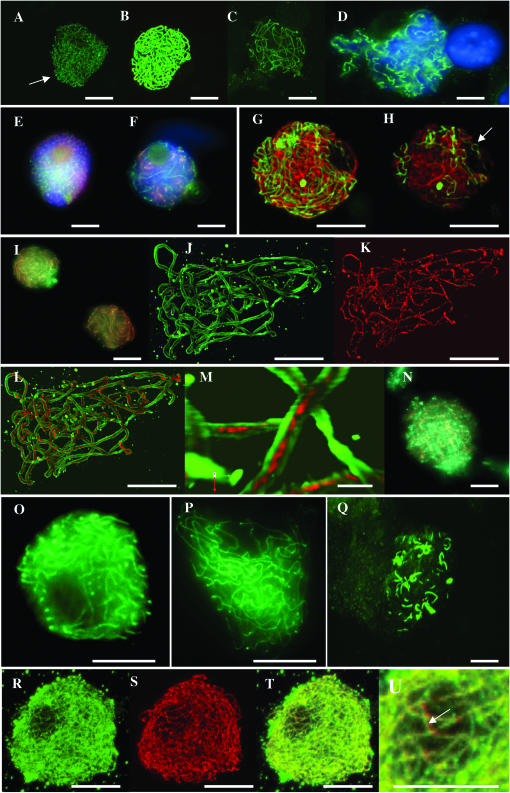
To explore the molecular basis for the disturbance of meiosis in this mutant, meiotic prophase cells of wild type and sy10 were probed with antibodies to two meiotic proteins associated with the SC. Asy1 protein of A. thaliana is associated with meiotic chromosome cores (Armstrong et al. 2002), and Zyp1 protein of A. thaliana is a component of the transverse filaments of the SC (Higgins et al. 2005). At leptotene in wild-type rye, Asy1 protein forms thin linear signals (Figure 3A). These probably represent unpaired chromosome cores or are in close apposition to axial elements since they form distinct bouquets at this stage. As meiotic prophase progresses to zygotene, the linear signals thicken commensurately with synapsis and chromosome condensation (Figure 3B). At pachytene (Figure 3C), the protein structures are shorter and appear to delimit the axes of bivalents. By diakinesis (Figure 3D), the protein is disorganized and adopts spiral conformations. Simultaneous probing of leptotene cells with antibodies to Asy1 and Zyp1 highlights separate linear structures (Figure 3E). During early synapsis separate linear structures can still be detected, irrespective of whether anti-Asy1 antibody is used at the same time with an antibody to the N terminus of Zyp1 (Figure 3F) or to the C terminus (Figure 3, G and H). The single optical section shown in Figure 3H confirms not only the stage (the presence of pairing forks), but also the clear physical separation of linear tracts of the two proteins. At late zygotene when synapsis is virtually complete, Asy1 and Zyp1 proteins are largely colocalized when viewed with conventional epifluorescence (Figure 3I). With the increased resolving power of confocal imagery and deconvolution of optical stacks, the two proteins can more correctly be described as coaligned at the later synaptic stages. Figure 3J is the separated channel of an optical stack through a pachytene cell and shows continuous, ribbon-like tracts of Asy1 protein. Each tract comprises two parallel linear structures of the protein separated by a fixed gap of similar width to the central region of the SC. Figure 3K shows unbroken, single linear tracts of Zyp1 protein in the same cell. Merging the two channels clearly demonstrates that Zyp1 protein occupies the space between the parallel linear structures of Asy1 (Figure 3, L and M). The coalignment of two Asy1 linear structures and one Zyp1 core into a tripartite linear structure is entirely consistent with their roles elaborated in A. thaliana. At diplotene, both Asy1 and Zyp1 proteins adopt spiral conformations as the SC is dismantled (Figure 3N). The same peculiarity of diplotene–diakinesis transformations of SCs in male meiosis of rye has been shown previously by electron microscopy (Fedotova et al. 1989).
Figure 3.—
Projections of optical sections through PMCs of wild type showing immunolocalization of Asy1 protein at (A) leptotene (bouquet indicated by arrow), (B) zygotene, and (C) pachytene. (D) A conventional fluorescence image of Asy1 protein at diakinesis with dual immunolocalization and conventional fluorescence imaging of anti-Asy1 (red) and anti-Zyp1N (green) antibodies in wild type at (E) leptotene and (F) zygotene. (G) Projection of optical sections through a PMC of wild type showing dual immunolocalization of anti-Asy1 (red) and anti-Zyp1C (green) antibodies. (H) Single section from the optical stack used in G, confirming both the stage (pairing fork indicated by arrow) and the physical separation of linear tracts of the two proteins. (I) Conventional fluorescence image of late zygotene in wild type, showing virtually complete colocalization of Asy1 (red) and Zyp1 (green) proteins. Projection of optical sections through a PMC at pachytene of wild type, showing dual immunolocalization of antibodies to Asy1 protein (J), Zyp1C protein (K), and merged channels (L). (M) Detail from L showing that the Zyp1 signal is clearly sandwiched between Asy1 at this stage. (N) Conventional fluorescence image showing that Asy1 and Zyp1 proteins adopt spiral conformations at diplotene in wild type. Immunolocalization of Asy1 protein (green) in the sy10 mutant at stages equivalent to zygotene (O), pachytene (P), and diakinesis (Q). Projection of optical sections through a PMC at midprophase in the sy10 mutant, showing dual immunolocalization of antibodies to Zyp1N (green), Asy1 (red), merged channels (T), and detail showing coaligned linear signals of the two proteins (arrow in U). Bars, 10 μm, except that of M, which represents 1 μm.
Immunolocalization of the two proteins in the mutant showed two interesting features. First, as deduced from observations by electron microscopy, the mutant builds normal axial cores (Figure 3O). At pachytene the protein fails to delimit pachytene bivalents as in wild type because of the widespread asynapsis (Figure 3P). Diakinesis is also unlike that in wild type, in that the protein does not form spiral structures (Figure 3Q). Second, although few SC segments are seen by electron microscopy, extensive linear tracts of Zyp1 and Asy1 are abundant in the nuclei (Figure 3, R and S, respectively) and coalign into bipartite ribbons (Figure 3, T and U). In other words, asynapsis in this mutant is not the direct result of failure of recruitment and association of Asy1 and Zyp1 proteins on chromosome cores.
DISCUSSION
Molecular assembly of bivalents in wild type:
The protein Asy1 of A. thaliana is associated with meiotic axes in this species and its relative Brassica oleracea (Armstrong et al. 2002), and it has been shown to have orthologs in yeast (Hollingsworth and Byers 1989), rice (Nonomura et al. 2004a,b, 2006), and most probably maize (Hamant et al. 2006). The protein Zyp1 of A. thaliana is a component of the transverse filaments of the SC (Higgins et al. 2005) and has been shown to have equivalent proteins in yeast (Sym et al. 1993), mammals (Heyting 1996), Caenorhabditis elegans (Couteau et al. 2004), and D. melanogaster (Page and Hawley 2004). Antibodies raised against these two proteins of A. thaliana were used in this study to monitor and compare the assembly of bivalents in wild type and its synaptic mutant sy10. First, both antibodies recognized strongly specific epitopes in rye, indicating a significant degree of structural similarity between the orthologous proteins, as also has been shown for Zyp1 protein of Triticum monococcum (Osman et al. 2006). Optical reconstruction of meiotic bivalents at pachytene, challenged by the two antibodies, showed that the spatial expression of the two proteins closely mirrored that in A. thaliana and B. oleracea. At this stage, continuous linear tracts of Zyp1 protein are flanked by continuous tracts of Asy1 protein, in keeping with their roles in the assembly of bivalents deduced from observations in A. thaliana. However, the temporal expression of the two proteins in rye appears to be different. In A. thaliana, foci of Asy1 protein appear at the onset of meiosis and then extend or fuse through chromosome condensation to form continuous linear signals by the end of leptotene. These tracts are maintained until the SC is dismantled at diplotene and diakinesis (Armstrong et al. 2002). The progression of assembly and retention of linear tracts of Asy1 protein appear similar in rye when its antibody is used on its own. However, simultaneous immunolocalization with both antibodies delimits separate tracts of both proteins at leptotene and zygotene. With the reasonable assumption that both proteins are nucleating on or near chromosome axes, the implication is that the tracts of Asy1 protein are discontinuous at these two stages. The discontinuity could simply be the result of prolonged loading of Asy1 protein due to the large size of the genome compared with A. thaliana. Alternatively, the discontinuity could be the result of a preselection of certain sequences only for incorporation into the SC. This could also be an adaptation to a large genome and has important implications for predisposition of sequences to recombination. Differences in expression of Asy1 protein have been noted in other organisms. In rice, the Asy1 ortholog PAIR2 loads early, before the onset of meiosis, and the signals extend at early meiotic prophase. However, during synapsis the signals become fragmented and faint (Nonomura et al. 2006). A similar loading pattern was observed in maize probed with Asy1 antibodies from Arabidopsis, which has continuous signals at early prophase, but, as synapsis proceeds, loses the signals (Hamant et al. 2006). Clearly, these species, together with rye, have adopted different strategies to assemble meiotic bivalents.
Foci of the protein Zyp1 have been shown in A. thaliana to form at pairing forks and linearize commensurately along bivalents with synapsis of homologs (Higgins et al. 2005). However, in rye, linear signals of Zyp1 appear at leptotene before synapsis has started. The inference is that monomers of Zyp1 can nucleate along the axes of unpaired meiotic chromosomes before synapsis and spatially separate from Asy1.
Aberrant chromosome interaction in the sy10 mutant:
The sy10 mutant is one of five nonallelic mutants of rye described to date showing indiscriminate synapsis (Fedotova et al. 1994; Sosnikhina et al. 1994a,b, 2001, 2002a,b). The sy10 mutation appears to not affect the timing or expression of Asy1 and Zyp1 proteins, so it is clearly not an allele of the ASY1 or ZYP1 orthologs of rye, despite some superficial phenotypic similarities with the mutants asy1 (Caryl et al. 2000) and zyp1 (Higgins et al. 2005). Both proteins form linear structures during zygotene, but these fail to assemble into coaligned ribbons at pachytene, which is characteristic of wild type. Interestingly, unsynapsed regions appear to comprise one linear signal each of the two proteins in coalignment at midmeiotic prophase, indicating that the asynaptic phenotype is not the direct result of failure of association of the two proteins per se. Clearly, the mutant phenotype is the result of a lesion in another gene, either responsible for building the tripartite structure of the SC or, more likely, connected with the initiation and progression of recombination to which synapsis is coupled. Preliminary experiments to assay levels of recombination by immunolocalization of Spo11 and Rad51/Dmc1 proteins in this mutant have so far proven inconclusive and are not reported here. Also, it is doubtful at present if antibodies to proteins responsible for late recombination events, such as anti-Mlh1, are going to be effective markers of recombination in this largely asynaptic and achiasmate mutant.
Electron microscopy of surface-spread meiocytes reveals that the sy10 mutant phenotype is clearly manifested during meiotic prophase: there is widespread asynapsis and exchanges of pairing partners, the latter indicating that heterologs are permitted to form SCs. It is not possible to discern a pattern in synapsis at this stage, since individual axial elements are difficult to track and chromosome identities are not known at this level. It is also impossible to say whether or not SCs themselves are effective in terms of supporting crossing over. However, inferences may be drawn about the nature of synapsis from the study of chromosome association at first metaphase. These data show that chromosome 4R forms a disproportionately high number of chiasmata at this stage compared with other chromosomes of the complement. Since the SC provides the platform for recombination events, the inference is that chromosome 4R had a higher propensity to form effective SCs compared with other chromosomes. This chromosome has a small amount of heterochromatin compared with other chromosomes of the set, as well as a pronounced chromatin extension in the form of tertiary constriction in its short arm. This unusual packing of chromatin could predispose this chromosome to the action of recombinogenic enzymes and result in its higher involvement in chiasmate and nonchiasmate associations compared with the other chromosomes. Chromosome 5R, which is also characterized by the presence of a weakly compacted region in its long arm, is involved in nonhomologous chiasmate associations only, which involve particularly this arm and the extended region in the short arm of 4R.
The data also show that nonhomologous chromosomes form chiasmate associations that are indistinguishable at the level of the light microscope from their homologous counterparts. If these represent true crossing over between nonhomologous DNA molecules, as in the zyp1 mutant of A. thaliana (Higgins et al. 2005), the inference is that some of the nonhomologous SCs at meiotic prophase are effective. It is not possible at present to determine if the promiscuity of chiasma formation is actually governed by an underlying DNA sequence homology between common repetitive sequences on nonhomologous chromosomes. Sticky, nonchiasmate associations may reflect the promiscuity of chromosome configurations at an earlier stage of meiosis. However, since they are largely random, it would appear that they have no functional significance in terms of rescuing achiasmate meiosis, as has been suggested for desynaptic (Orellana and Giraldez 1983) and inbred rye (Cermeno et al. 1984).
The presence of univalents in sy10 mutant and the low number of homologous bivalents correspond to elevated levels of ectopic recombination and reduced levels of allelic recombination observed in the mutant hop2 characterized by indiscriminate synapsis (Leu et al. 1998). It is also thought that the success of pairing depends on the bringing together of chromosomes into close proximity (bouquet formation), which leads to close contacts between telomeres and between chromosomes in the nucleus, and this process facilitates homology testing (Zickler 2006). In the case of hop2 of S. cerevisiae, Leu et al. (1998) exclude the possibility that the hop2 protein participates in telomere-dependent pairing, since immunodetection indicates that this protein is not concentrated at chromosome ends. Scrutiny of early meiotic prophase shows that the telomeres in sy10 form a distinct bouquet. It differs in this respect from the sy1 mutant of rye (Mikhailova et al. 2001) and the phs1 mutant of maize (Pawlowski et al. 2004), which have some disturbance in nuclear architecture at this stage of meiosis. Therefore it can be proposed that in the mutant sy10 either homologous sequences are not recognized and synapsis occurs at random between nonhomologous chromosomes or recognition involves all regions in allelic and ectopic positions, but there is no correction in the progress of meiosis, as occurs in the wild type of certain species (Zickler and Kleckner 1999). The early meiotic pairing may include the formation of nonstable connections (Kleckner 2006). In this case, nonstable associations between ectopic homologous repeats may be resolved as meiosis progresses. It is possible that the activity of SY10 reveals itself especially at this stage and that its function is to ensure strict homology of pairing. The corollary is that SY10 functions at the levels of pairing, testing homology, and stabilizing interhomologous bonds.
In conclusion, this study demonstrates that the mutant allele of SY10 is responsible for failure of the integrity of synapsis and chiasma formation. However, this phenotype is not the result of compromising the spatial or temporal expression of two proteins integrally involved in the molecular assembly of meiotic chromosomes. Rather, this study points to an aberration in the recombination machinery to which other events of meiotic prophase are coupled. The study also highlights some important differences in the recruitment of orthologs in rye compared with A. thaliana and emphasizes the necessity of integrating our knowledge of meiosis from distantly related organisms.
Acknowledgments
We are indebted to Chris Franklin, Gareth Jones, and Sue Armstrong (Birmingham) for supplying the antibodies used in this study. We are also grateful to Denise Zickler (Paris) and Zac Cande (Berkeley) for their most helpful comments on the manuscript, and to Vyacheslav Lovtsyus for database programming. We thank the Leverhulme Trust (Emeritus Fellowship and Research Grant to R.N.J.), the Thomas and Elizabeth Williams Fund (D.P.), the Russian Foundation for Basic Research (grants 03-04-48887 and 06-04-48419a), Civilian Research and Development Foundation (grant ST-012-0), and the Biotechnology and Biological Sciences Research Council for financial support. This work was also funded within the Program of the Presidium of the Russian Academy of Sciences “Biodiversity and Dynamics of the Gene Pool” and supported by the President of the Russian Federation (grant in support of Leading Scientific Schools 2214.2003.4).
References
- Abbo, M. W., R. P. Dunford, T. Foote, S. M. Reader, R. B. Flavell et al., 1995. Organisation of retroelement and stem-loop repeat families in the genomes and nuclei of cereals. Chromosome Res. 3: 5–15. [DOI] [PubMed] [Google Scholar]
- Alkhimova, A. G., J. S. Heslop-Harrison, A. I. Shchapova and A. V. Vershinin, 1999. Rye chromosome variability in wheat-rye addition and substitution lines. Chromosome Res. 7: 205–212. [DOI] [PubMed] [Google Scholar]
- Alkhimova, O. G., N. A. Mazurok, T. A. Potapova, S. M. Zakian, J. S. Heslop-Harrison et al., 2004. Diverse patterns of the tandem repeats organization in rye chromosomes. Chromosoma 113: 42–52. [DOI] [PubMed] [Google Scholar]
- Anderson, L. K., S. M. Stack, R. J. Todd and R. P. Ellis, 1994. A monoclonal antibody to lateral element proteins in synaptonemal complexes of Lilium longiflorum. Chromosoma 103: 357–367. [DOI] [PubMed] [Google Scholar]
- Aragon-Alcaide, L., T. Miller, T. Schwarzacher, S. Reader and G. Moore, 1996. A cereal centromeric sequence. Chromosoma 10: 261–268. [DOI] [PubMed] [Google Scholar]
- Armstrong, S. J., A. P. Caryl, G. H. Jones and F. C. H. Franklin, 2002. Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J. Cell Sci. 115: 3645–3655. [DOI] [PubMed] [Google Scholar]
- Bishop, D. K., and D. Zickler, 2004. Early decision: meiotic crossover interference prior to stable strand exchange and synapsis. Cell 117: 9–15. [DOI] [PubMed] [Google Scholar]
- Bogdanov, Y. F., 2003. Variation and evolution of meiosis. Russ. J. Genet. 39: 453–473. [PubMed] [Google Scholar]
- Caryl, A. P., S. J. Armstrong, G. H. Jones and F. C. H. Franklin, 2000. A homologue of the yeast HOP1 gene is inactivated in Arabidopsis meiotic mutant asy1. Chromosoma 109: 62–71. [DOI] [PubMed] [Google Scholar]
- Cermeno, M. C., J. Orellana and J. R. Lacadena, 1984. Evidence of nonchiasmate bonds at metaphase I in inbred rye. Can. J. Genet. Cytol. 26: 409–414. [Google Scholar]
- Chatterjee, R., and G. Jenkins, 1993. Meiotic chromosome interactions in inbred autotetraploid rye (Secale cereale L.). Genome 36: 131–138. [DOI] [PubMed] [Google Scholar]
- Couteau, F., K. Nabeshima, A. Villeneuve and M. Zetka, 2004. A component of C. elegans meiotic chromosome axes at the interface of homolog alignment, synapsis, nuclear reorganization, and recombination. Curr. Biol. 14: 585–592. [DOI] [PubMed] [Google Scholar]
- Cuadrado, A., and N. Jouve, 2002. Evolutionary trends of different repetitive DNA sequences during speciation in the genus Secale. J. Hered. 93: 339–345. [DOI] [PubMed] [Google Scholar]
- Cuadrado, A., C. Ceoloni and N. Jouve, 1995. Variation in highly repetitive DNA composition of heterochromatin in rye studied by fluorescence in situ hybridization. Genome 38: 1061–1069. [DOI] [PubMed] [Google Scholar]
- Devos, K. M., and M. D. Gale, 2000. Genome relationships: the grass model in current research. Plant Cell 12: 637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos, K. M., M. D. Atkinson, C. N. Chinoy, H. A. Francis, R. L. Harcourt et al., 1993. Chromosomal rearrangements in the rye genome relative to that of wheat. Theor. Appl. Genet. 85: 673–680. [DOI] [PubMed] [Google Scholar]
- Fedotova, Y. S., O. L. Kolomiets and Y. F. Bogdanov, 1989. Synaptonemal complex transformations in rye microsporocytes at the diplotene stage of meiosis. Genome 32: 816–823. [Google Scholar]
- Fedotova, Y. S., Y. F. Bogdanov, S. A. Gadzhiyeva, S. P. Sosnikhina, V. G. Smirnov et al., 1994. Meiotic mutants of rye Secale cereale L. II. The nonhomologous synapsis in desynaptic mutants sy7 and sy10. Theor. Applied Genet. 88: 1029–1036. [DOI] [PubMed] [Google Scholar]
- Gerlach, W. L., and T. A. Dyer, 1980. Sequence organization of the repeating units in the nucleus of wheat which contain 5S rRNA genes. Nucleic Acids Res. 11: 4851–4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerton, J. L., and R. S. Hawley, 2005. Homologous chromosome interactions in meiosis: diversity amidst conservation. Nat. Rev. Genet. 6: 477–487. [DOI] [PubMed] [Google Scholar]
- Hajdera, I., D. Siwinska, R. Hasterok and I. Maluszynska, 2003. Molecular cytogenetic analysis of genome structure in Lupinus angustifolius and Lupinus cosentinii. Theor. Appl. Genet. 107: 988–996. [DOI] [PubMed] [Google Scholar]
- Hamant, O., H. Ma and W. Z. Cande, 2006. Genetics of meiotic prophase I in plants. Annu. Rev. Plant Biol. 57: 267–302. [DOI] [PubMed] [Google Scholar]
- Heyting, C., 1996. Synaptonemal complexes: structure and function. Curr. Opin. Cell Biol. 8: 389–396. [DOI] [PubMed] [Google Scholar]
- Heyting, C., A. J. Dietrich, J. H. de Jong and E. Hartsuiker, 1994. Immunocytochemical techniques applied to meiotic chromosomes. Methods Mol. Biol. 29: 287–301. [DOI] [PubMed] [Google Scholar]
- Higgins, J. D., E. Sanchez-Moran, S. J. Armstrong, G. H. Jones and F. C. H. Franklin, 2005. The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev. 19: 2488–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth, N. M., and B. Byers, 1989. HOP1: a yeast meiotic pairing gene. Genetics 121: 445–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins, G., E. I. Mikhailova, T. Langdon, O. A. Tikholiz, S. P. Sosnikhina et al., 2005. Strategies for the study of meiosis in rye. Cytogenet. Genome Res. 109: 221–227. [DOI] [PubMed] [Google Scholar]
- Kleckner, N., 2006. Chiasma formation: chromati/axis interplay and the role(s) of the synaptonemal complex. Chromosoma 115: 175–194. [DOI] [PubMed] [Google Scholar]
- Langdon, T., C. Seago, M. Mende, M. Leggett, H. Thomas et al., 2000. Retrotransposon evolution in diverse plant genomes. Genetics 156: 313–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu, J.-Y., P. R. Chua and G. S. Roeder, 1998. The meiosis-specific Hop2 protein of S. cerevisiae ensures synapsis between homologous chromosomes. Cell 94: 375–386. [DOI] [PubMed] [Google Scholar]
- Mikhailova, E. I., S. P. Sosnikhina and T. G. Michurina, 1993. Identification of rye chromosomes by means of conjugation test in meiosis. Genetika 29: 978–989. [Google Scholar]
- Mikhailova, E. I., S. P. Sosnikhina, G. A. Kirillova, O. A. Tikholiz, V. G. Smirnov et al., 2001. Nuclear dispositions of subtelomeric and percentromeric chromosomal domains during meiosis in asynaptic mutants of rye (Secale cereale L.). J. Cell Sci. 114: 1875–1882. [DOI] [PubMed] [Google Scholar]
- Nonomura, K.-I., M. Nakano, K. Murata, K. Miyoshi, M. Eiguchi et al., 2004. a An insertional mutation in the rice PAIR2 gene, the ortholog of Arabidopsis ASY1, results in a defect in homologous chromosome pairing during meiosis. Mol. Genet. Genomics 271: 121–129. [DOI] [PubMed] [Google Scholar]
- Nonomura, K. I., M. Nakano, T. Fukuda, M. Eiguchi, A. Miyao et al., 2004. b The novel gene HOMOLOGOUS PAIRING IN RICE MEIOSIS1 of rice encodes a putative coiled-coil protein required for homologous chromosome pairing in meiosis. Plant Cell 16: 1008–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura, K.-I., M. Nakano, M. Eiguchi, T. Suzuki and N. Kurata, 2006. PAIR2 is essential for homologous chromosome synapsis in rice meiosis I. J. Cell Sci. 119: 217–225. [DOI] [PubMed] [Google Scholar]
- Orellana, J., and R. Giraldez, 1983. Metaphase I bound arms and crossing over frequency in rye III. Non-chiasmate bonds in desynaptic plants. Heredity 51: 383–394. [Google Scholar]
- Osman, K., E. Sanchez-Moran, J. D. Higgins, G. H. Jones and F. C. H. Franklin, 2006. Chromosome synapsis in Arabidopsis: analysis of the transverse filament protein ZYP1 reveals novel functions for the synaptonemal complex. Chromosoma 115: 212–219. [DOI] [PubMed] [Google Scholar]
- Page, S. L., and R. S. Hawley, 2004. The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 20: 525–558. [DOI] [PubMed] [Google Scholar]
- Pawlowski, W. P., I. N. Golubovskaya, L. Timofejeva, R. B. Meeley, W. F. Sheridan et al., 2004. Coordination of meiotic recombination, pairing and synapsis by PHS1. Science 303: 89–92. [DOI] [PubMed] [Google Scholar]
- Rees, H., 1955. Genotypic control of chromosome behaviour in rye I. Inbred lines. Heredity 9: 93–116. [Google Scholar]
- Rees, H., and J. B. Thompson, 1956. Genotypic control of chromosome behaviour in rye III. Chiasmata frequency in homozygotes and heterozygotes. Heredity 10: 409–424. [Google Scholar]
- Schulman, A. H., P. K. Gupta and R. K. Varshney, 2004. Organization of retrotransposons and microsatellites in cereal genomes., pp. 83–118 in Cereal Genomics, edited by P. K. Gupta and R. K. Varshney. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- Shaw, P., and G. Moore, 1998. Meiosis: vive la difference! Curr. Opin. Plant Biol. 1: 458–462. [DOI] [PubMed] [Google Scholar]
- Sosnikhina, S. P., Y. S. Fedotova, V. G. Smirnov, E. I. Mikhailova and Y. F. Bogdanov, 1994. a The study of genetic control of meiosis in rye. Russ. J. Genet. 30: 909–920. [Google Scholar]
- Sosnikhina, S. P., V. G. Smirnov, E. I. Mikhailova and L. F. Egorova, 1994. b Homologous synapsis distortion in meiotic mutants of diploid rye. Russ. J. Genet. 30: 431–436. [Google Scholar]
- Sosnikhina, S. P., G. A. Kirillova, E. I. Mikhailova, O. A. Tikholiz, V. G. Smirnov et al., 2001. Genetic control of chromosome synapsis at meiosis in rye Secale cereale L.: the sy19 gene controlling heterologous synapsis. Russ. J. Genet. 37: 71–79. [Google Scholar]
- Sosnikhina, S. P., G. A. Kirillova, O. A. Tikholiz, E. I. Mikhailova, S. N. Priyatkina et al., 2002. a Genetic analysis of mutation sy2 which causes nonhomologous meiotic synapsis in chromosomes of diploid rye Secale cereale L. Russ. J. Genet. 38: 269–276. [PubMed] [Google Scholar]
- Sosnikhina, S. P., G. A. Kirillova, O. A. Tikholiz, E. I. Mikhailova, V. G. Smirnov et al., 2002. b The expression of mutation sy2 causing nonhomologous synapsis in meiosis of diploid rye Secale cereale L. Russ. J. Genet. 38: 156–164. [PubMed] [Google Scholar]
- Sosnikhina, S. P., E. I. Mikhailova, O. A. Tikholiz, S. N. Priyatkina, V. G. Smirnov et al., 2005. Meiotic mutations in rye Secale cereale L. Cytogenet. Genome Res. 109: 215–220. [DOI] [PubMed] [Google Scholar]
- Sybenga, J., 1983. Rye chromosome nomenclature and homoeology relationships. Z. Pflanzenzuechtung 90: 297–307. [Google Scholar]
- Sym, M., J. A. Engebrecht and G. S. Roeder, 1993. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell 72: 365–378. [DOI] [PubMed] [Google Scholar]
- Terasawa, M., A. Shinohara, Y. Hotta, H. Ogawa and T. Ogawa, 1995. Localization of RecA-like recombination proteins on chromosomes of the lily at various meiotic stages. Genes Dev. 9: 925–934. [DOI] [PubMed] [Google Scholar]
- Tikhonovich, I. A., and T. S. Fadeyeva, 1976. The study of heterochromatin in karyotypes of the forms of genetic collections of rye. Genetika 12: 5–14. [Google Scholar]
- Tikhonovich, I. A., A. G. Dubovskaya and S. P. Sosnikhina, 1987. The use of heterochromatin markers of chromosomes in cytogenetic studies in rye (Secale cereale L.). Part 1. The behaviour of chromosomes 1 (7R) and 5 (4R) in meiosis. Genetika 23: 838–844. [Google Scholar]
- Unfried, I., and P. Gruendler, 1990. Nucleotide sequence of the 5.8S and 25S rRNA genes and the internal transcribed spacers from Arabidopsis thaliana. Nucleic Acids Res. 18: 4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney, R. K., V. Korzun and A. Börner (Editors), 2004. Molecular Maps in Cereals: Methodology and Progress. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- Vershinin, A. V., T. Schwarzacher and J. S. Heslop-Harrison, 1995. The large-scale genomic organization of repetitive DNA families at telomeres of rye chromosomes. Plant Cell 7: 1823–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler, D., 2006. From early homologue recognition to synaptonemal complex formation. Chromosoma 115: 158–174. [DOI] [PubMed] [Google Scholar]
- Zickler, D., and N. Kleckner, 1998. The leptotene-zygotene transition of meiosis. Annu. Rev. Genet. 32: 619–697. [DOI] [PubMed] [Google Scholar]
- Zickler, D., and N. Kleckner, 1999. Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 33: 603–754. [DOI] [PubMed] [Google Scholar]