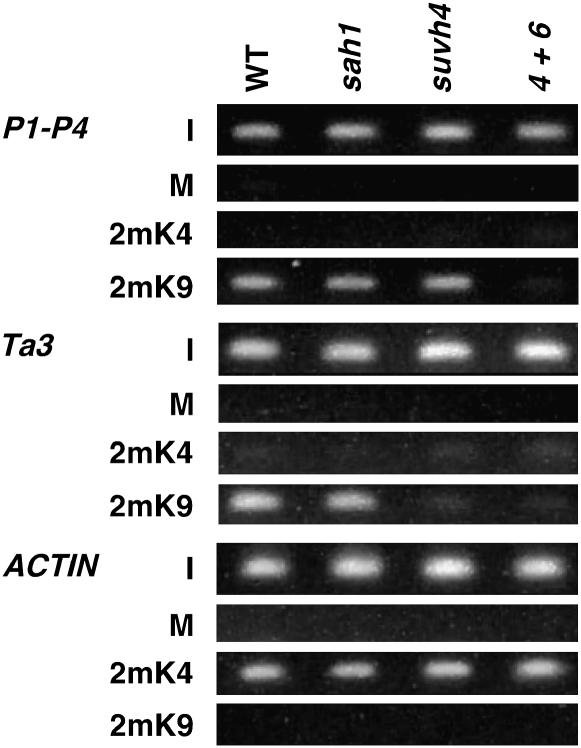
Figure 7.—
The sah1L459F mutation does not deplete H3 2mK9 or H3 2mK4 from modified regions. ChIP analysis of the indicated mutants is shown. WT is Ws pai1, sah1 is Ws pai1 sah1L459F, suvh4 is Ws pai1 suvh4, and 4+6 is pai1 suvh4 suvh6 (Ebbs et al. 2005). Primer sets specific for each locus were used to amplify PCR products from total-input chromatin (I), no antibody mock precipitation control (M), chromatin immunoprecipitated with H3 anti-dimethyl K9 antibodies (2mK9), or chromatin immunoprecipitated with H3 anti-dimethyl K4 antibodies (2mK4) from the indicated mutants. GelStar-stained PCR products are shown. P1–P4 indicates the PAI1–PAI4 inverted repeat.