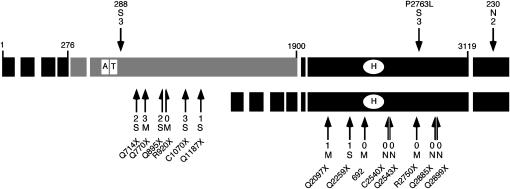
Figure 3.—
Summary of the Su(var)2-HP2 alleles by type and location. Each exon is indicated by a large rectangle; the shaded exons are the HP2-L-specific exons, and the solid exons are common to both HP2-L and HP2-S (HP2-L, top line; HP2-S, bottom line). The small numbers at the ends of some of the exons of the larger isoform indicate the number of the amino acid found at the exon boundary. The open rectangles labeled “A” and “T” indicate the location of the two AT hooks found within HP2-L. The open oval labeled “H” indicates the position of the HP1-binding domain (Stephens et al. 2005). The location of each mutation described is indicated with a solid arrow. Arrows above the larger isoform and pointing downward indicate the position of the three missense alleles. The arrows and half-arrows below the isoforms and pointing upward indicate the locations of the truncation alleles. The number at the end of the arrow indicates the level of suppression of variegation, from near complete (3) to no (0) loss of silencing (see Figure 5). The letter above or below the Su(var) number indicates the extent of mitotic abnormalities: S, very frequent abnormalities; M, medium level of abnormalities; N, no abnormalities (see Table 2). The amino acid change for allele 288 is T588I, the amino acid change for 230 is N3220I, and allele 692 is a single-base insertion of an A, which produces a frameshift at amino acid 2370. The amino acid change for all other alleles is indicated by the allele name.