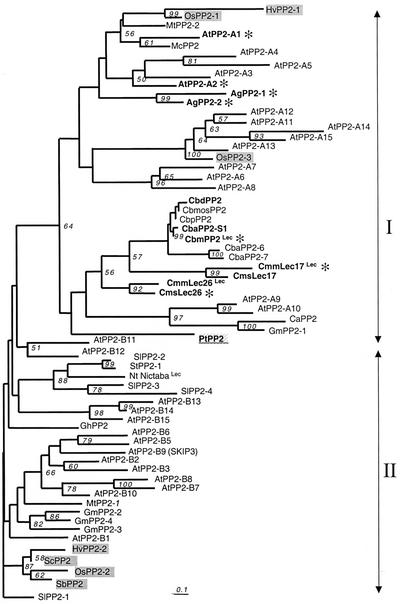
Figure 6.
Phylogenetic relationship of PP2-like proteins. Amino acid sequences of the PP2 conserved domain (motifs A–D) were aligned using ClustalW (Thompson et al., 1994). After corrections for an high rate of substitution using the Kimura 2-parameters distance option included in ClustalW, a neighbor-joining tree (Saitu and Nei, 1987) depicting relationships among the PP2-like proteins was constructed by using the aligned amino acid sequences described in Table III. Two sequences, PpPP2 and AtPP2-B4, that showed only partial alignments were not included in the tree. The protein sequence deduced from a P. taeda EST (PtPP2) was included in the phylogenetic analysis represented as an unrooted tree. All branches are drawn to scale as indicated by the scale bar (bar = substitution/site rate of 0.1%), and their length indicates the level of divergence among sequences. Only percentages of bootstrap values supported by more than 50% of the 1,000 replicates are indicated above nodes. These sequences can be assembled into two main groups (arrows). For each protein, the prefix indicates the species from which it is derived. GenBank accession numbers and acronyms are detailed in Table III. Proteins from Arabidopsis, cucurbits, and celery that were more extensively described in this work are indicated in bold letters. Monocot proteins are indicated on a gray background. The gymnosperm protein PtPP2 is underlined on a gray background. Lec, Proteins for which GlcNAc-binding activity was demonstrated. A gray asterisk (*) indicates proteins corresponding to genes expressed in the companion cell-sieve element complex.