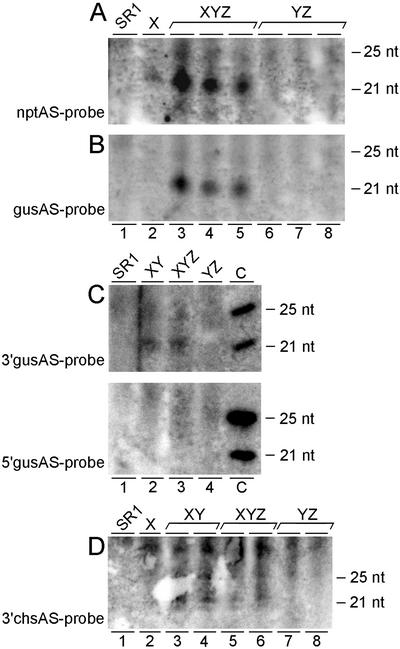
Figure 3.
Detection of small RNAs. Low-Mr RNA fractions were isolated from leaf tissue of mature non-flowering tobacco plants, separated on polyacrylamide gels, blotted onto Hybond N+ membranes, and hybridized with hydrolyzed antisense riboprobes: nptII (full-length nptII-coding sequence; A), gus (full-length gus-coding sequence; B), 3′gus (most 3′ 800 bp of the gus-coding sequence; C, top panel), 5′gus (most 5′ 800 bp of the gus-coding sequence; C, bottom panel), and 3′chs (3′chs-UTR sequence; D). DNA oligomers were used as size controls (size indicated in nucleotides). Each numbered lane contains the low-Mr RNA fraction of another tobacco plant of the genotype indicated on top. For the wild-type tobacco SR1 and the normal gus-expressing hybrid plants YZ, no specific signal could be detected with either of the probes. A, Locus X-containing plants (X and XYZ) gave rise to small nptII-specific RNAs of approximately 22 nucleotides. B, The XYZ plants showing gus silencing accumulated approximately 22-nucleotide small gus-specific RNAs. C, In gus-silenced XY and XYZ plants, these small RNAs corresponding to the 5′gus probe do not accumulate to detectable levels (bottom panel), whereas those corresponding to the 3′gus probe give a detectable signal upon identical exposure time (top panel). C, The lanes indicated by c (controls) contain 3.3 pmol of 21-nucleotide long DNA and 2.5 pmol of 25-nucleotide long DNA with a GC content of 72% and 71.4%, respectively, and are shown for comparison of probe quality and quantity in top and bottom panel; the control DNA oligonucleotides in the upper and lower panels are 100% complementary to a stretch in the 5′gus probe and 3′gus probe, respectively. D, Approximately 22-nucleotide small 3′chs-specific RNAs were, although expected for all X-containing samples, only detected in XY and XYZ samples.