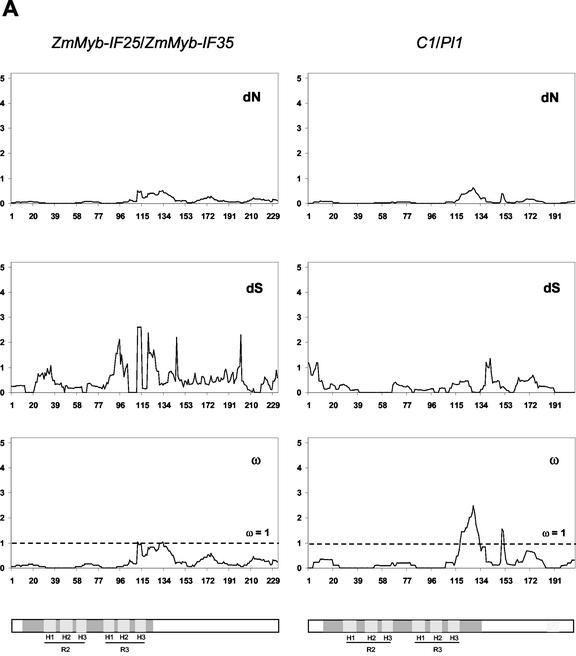
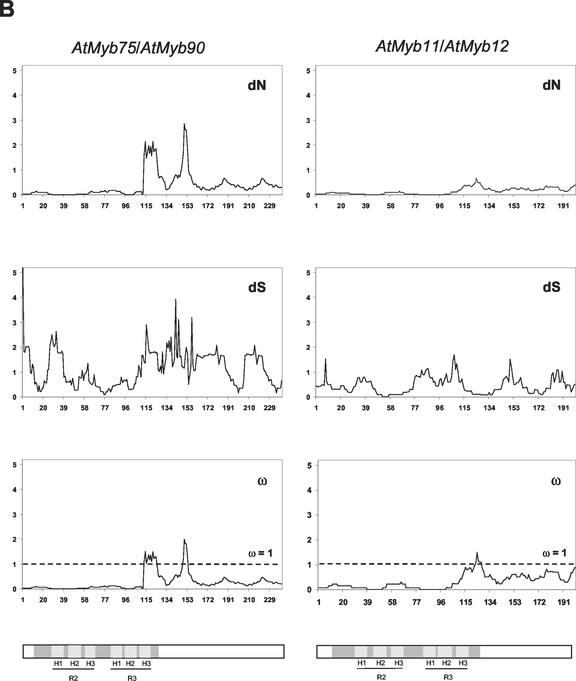
Figure 3.
Sliding window analysis of duplicated pairs of R2R3 Myb genes. Numbers of synonymous and non-synonymous substitutions in 15 codon windows were estimated using the YN00 model of sequence evolution (constraining nuisance parameters as described in “Materials and Methods”). Bars in gray indicate the three α-helices in MYB repeat 1 (H1, 22–33 amino acids; H2, 36–45 amino acids; and H3, 51–60 amino acids) and MYB repeat 2 (H1, 75–86 amino acids; H2, 88–97 amino acids; and H3, 102–111 amino acids), where the second and third helices form a helix-turn-helix (HTH) structure when bound to DNA. The TAD in ZmMyb-C1/ZmMyb-Pl1 is shown in light gray.