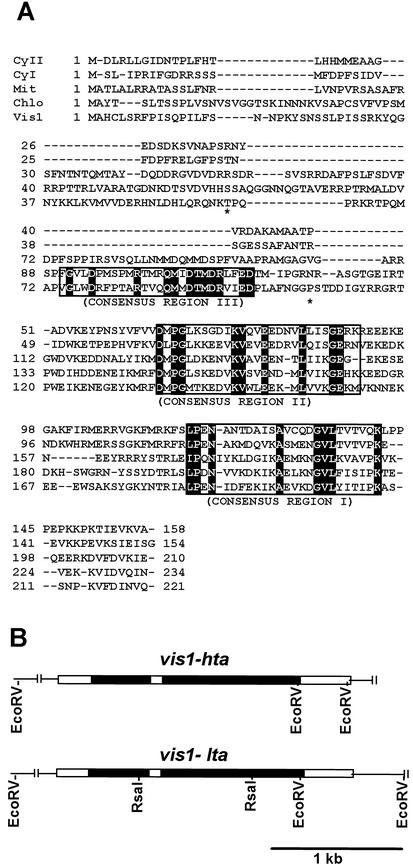
Figure 1.
Alignment of the deduced amino acid sequences of VIS1 with other tomato sHSPs (A) and genomic structures of vis1-hta and vis1-lta (B). A, Multiple sequence alignment was performed with ClustalX (Thompson et al., 1997) and manually edited. The three consensus regions in sHSPs are boxed, and identical amino acid residues are highlighted. The asterisks indicate the two residues, Thr61 and Pro107 in VIS1-HTA, that are replaced with Ala in VIS1-LTA. Dashes indicate gaps inserted to improve the alignment. Shown sHSPs are the CyI (cytoplasmic class I; accession no. CAA39603), CyII (cytoplasmic class II; accession no. AAC14577), Mit (mitochondrial; accession no. BAA32547), Chlo (chloroplastic pTOM111; accession no. AAB49626), and Vis1 (VIS1-HTA, accession no. AY128101). B, Shown are the two additional RsaI sites, one in each intron of vis1-lta (accession no. AY128102) but absent in vis1-hta (accession no. AY128101), that were used for developing a PCR-based assay for each allele. The black and white boxes represent introns and exons, respectively.