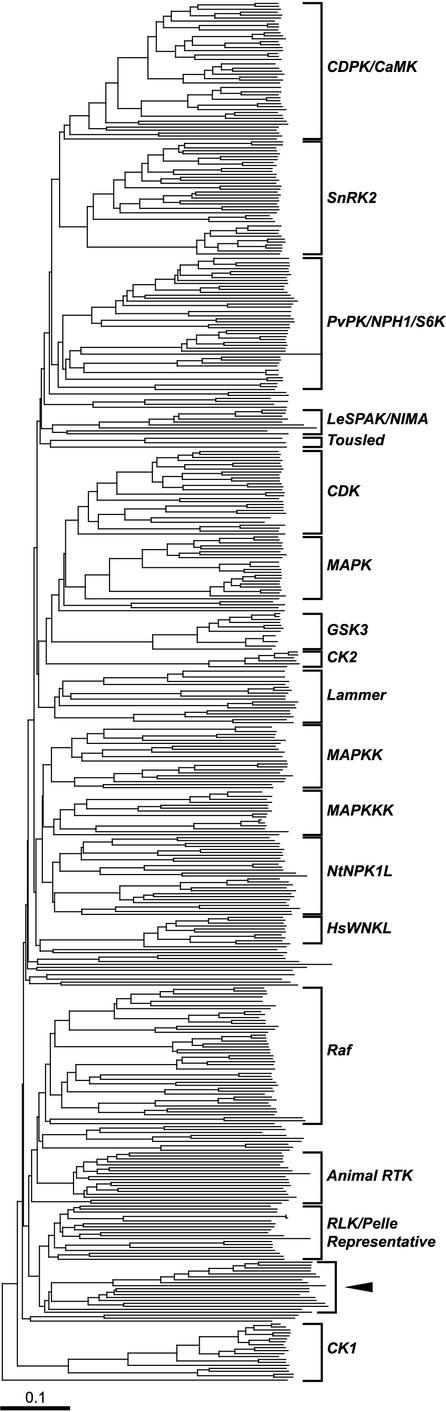
Figure 1.
Relationships among Arabidopsis protein kinases. The kinase sequences were identified from the Arabidopsis genome, and an alignment was generated using the kinase domain amino acid sequence. The phylogeny was inferred from the alignment using the neighbor-joining method with 100 bootstrap replicates (bootstrap values shown in Supplement B). The tentative kinase families were assigned based on nodes with more than 30% bootstrap support. The family names were given based on known sequences in each family. In the context of all kinases, representative RLK/Pelle members all fall into a monophyletic group as predicted. Interestingly, a group of kinases is found to form a sister group to the RLK/Pelle family (arrowhead a). In addition to RLKs in the RLK/Pelle family, the IRE1-like genes are the only other receptor kinases found in Arabidopsis (arrowhead b). The IRE1 family genes are not the closest relatives to RLK/Pelle members, suggesting that the receptor kinase configurations have arisen independently between these two kinase families. CDPK, Calcium-dependent protein kinase; CaMK, calmodulin-dependent kinase; SnRK2, SNF-related protein kinase 2; PvPK, bean protein kinase; NPH1, non-phototropic hypocotyl 1; S6K, ribosomal protein S6 kinase; LeSPAK, tomato self-pruning-associated kinase; NIMA, never in mitosis A; CDK, cyclin-dependent kinase; GSK3, glycogen synthase kinase 3; CK2, casein kinase 2; NtNPK1L, Nicotiana protein kinase 1 like; HsWNKL, human with no Lys kinase-like; RTK, receptor Tyr kinase; MAPK, mitogen-associated protein kinase; MAPKK, MAPK kinase; MAPKKK, MAPKK kinase.