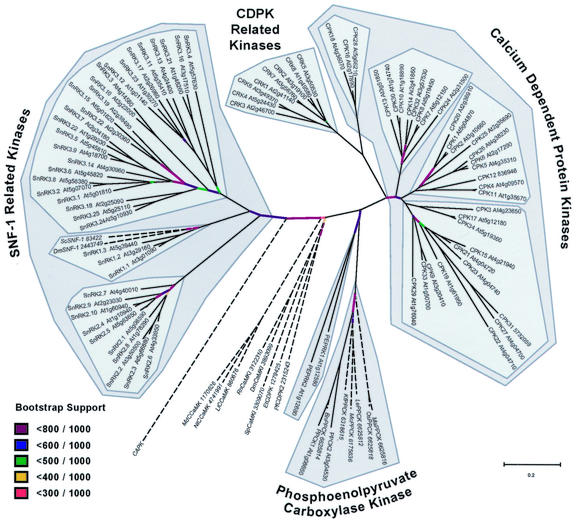
Figure 2.
Sequence tree for the Arabidopsis CDPK/SnRK superfamily based on alignment of kinase catalytic domains. The five kinase groups (CDPK, CRK, PPCK, PEPRK, and SnRK) are delimited by dark gray blocks. Subgroups within the CDPK and SnRK groups are shown as lighter shaded blocks. Non-Arabidopsis sequences included as outgroups are followed by sequence identifiers and are shown in italics and with dashed lines. CAPK denotes bovine cAMP-dependent protein kinase. Abbreviations for other organisms are: Sc, Brewer's yeast; Dm, fruitfly (Drosophila melanogaster); Md, apple (Malus domestica); Nt, tobacco (Nicotiana tabacum); Ll, Lilium longiflorum; Rn, Rattus norvegicus; Sp, fission yeast (Schizosac-charomyces pombe); Et, Eimeria tenella (apicomplexan); Pf, Plasmodium falciparum (apicomplexan); Bn, canola (Brassica napus); Kf, Kalanchoe fedtschenkoi; Mc, common ice plant (Mesembryanthemum crystallinum); Le, tomato (Lycopersicon esculentum); Os, rice (Oryza sativa); and Ma, Musa acuminata. Bootstrap supports for specific branches are shown in color. Noncolored branches are supported by greater than 800 trees in 1,000 bootstrap trials.