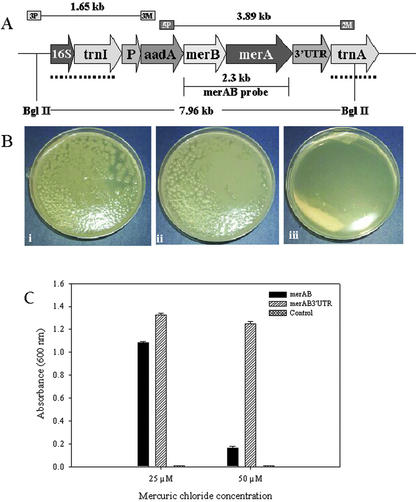
Figure 1.
Bacterial bioassay. A, Schematic representation of the transformed chloroplast genome: The map shows the transgenic chloroplast genome containing the pLDR-MerAB-3′-UTR construct. The site-specific integration between trnI and trnA chloroplast genes is shown by the dotted line, specifying the homologous recombination sequences in the pLDR-MerAB-3′-UTR and pLDR-MerAB. Landing sites for the 3P/3M and 5P/2M primer pairs used in PCR confirmation of integration, and expected sizes of products are shown. BglII restriction digestion sites and the merAB probe used in the Southern-blot analyses are shown. A fragment of 7.96 kb should be produced after restriction digestion of the transgenic chloroplast genome. B, Transformed E. coli grown in 100 μm HgCl2. i, Transformed E. coli cells containing the vectors pLDR-MerAB; ii, pLDR-MerAB-3′-UTR grown in Luria-Bertani at 100 μm HgCl2; iii, untransformed control (E. coli). C, Effect of mercuric chloride on E. coli cell proliferation. The transgenic clone pLDR-MerAB and pLDR-MerAB-3′-UTR and the control E. coli cells were grown on liquid Luria-Bertani medium with 25 and 50 μm of HgCl2 for 24 h at 37°C. The A600 was measured.