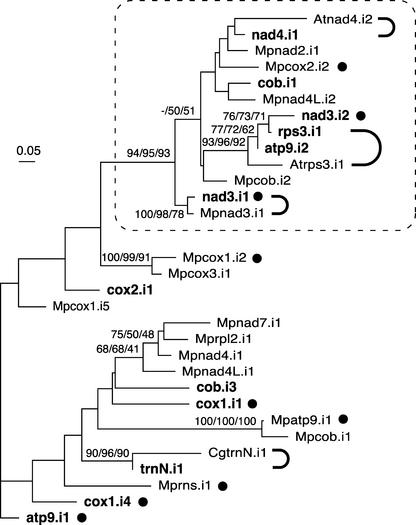
Figure 7.
Phylogenetic Analysis of Group-IIA Mitochondrial Introns from Chara and Marchantia and of Selected Introns from Chaetosphaeridium and Arabidopsis.
All Chara and Marchantia introns that were identified as belonging to group IIA were analyzed, whereas only those of Chaetosphaeridium and Arabidopsis that share insertion sites with their Chara counterparts were examined. Introns originating from Chara are indicated in boldface; Marchantia, Chaetosphaeridium, and Arabidopsis introns are distinguished by the prefixes Mp, Cg, and At, respectively. The introns whose names are followed by closed circles contain ORFs. The introns that are connected by thick, curved lines share common insertion sites. The nucleotide data set (122 sites corresponding to unambiguously aligned regions of the intron core) was analyzed using distance, maximum parsimony, and maximum likelihood methods. The distance and maximum likelihood analyses were performed using the Hasegawa-Kishino-Yano model and a uniform rate of substitutions across sites. The unrooted maximum likelihood tree is shown. Bootstrap values obtained in distance, maximum parsimony, and maximum likelihood analyses after 100 replications are indicated above the nodes in the top, middle, and bottom positions, respectively; only values of >50% are shown. A highly supported clade containing 13 introns is framed.