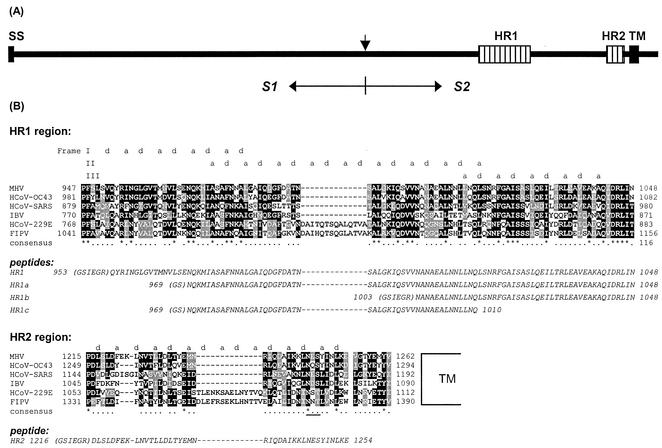
FIG. 1.
(A) Schematic representation of the coronavirus MHV-A59 spike protein structure. The glycoprotein has an N-terminal signal sequence (SS) and a transmembrane domain (TM) close to the C terminus. The protein is proteolytically cleaved (vertical arrow) into an S1 and an S2 subunit, which are noncovalently linked. S2 contains two HR regions (hatched bars), HR1 and HR2, as indicated. (B) Sequence alignment of HR1 and HR2 domains of MHV-A59 with those of HCoV-OC43, HCoV-229E, FIPV strain 79-1146, infectious bronchitis virus strain Beaudette (IBV), and the newly identified HCoV-SARS (strain TOR2). HCoV-229E and FIPV, MHV-A59 and HCoV-OC43, and IBV are representatives of groups 1, 2, and 3, respectively—the three coronavirus subgroups (59). Dark shading marks sequence identity, while lighter shading represents sequence similarity. The alignment shows a remarkable insertion of exactly two HRs (14 aa) in both HR1 and HR2 of HCV-229E and FIPV, a characteristic of all group 1 viruses. The predicted hydrophobic HR a and d residues are indicated above the sequence. The frame shifts in the predicted HRs in HR1 are caused by a stutter (51). The asterisks indicate conserved residues, and the dots represent similar residues. The amino acid sequences of the peptides HR1, HR1a, HR1b, HR1c, and HR2 used in this study are presented in italics below the alignments. N-terminal residues derived from the proteolytic cleavage site of the GST fusion protein are in parentheses. A conserved N-glycosylation sequence in the HR2 region is underlined.