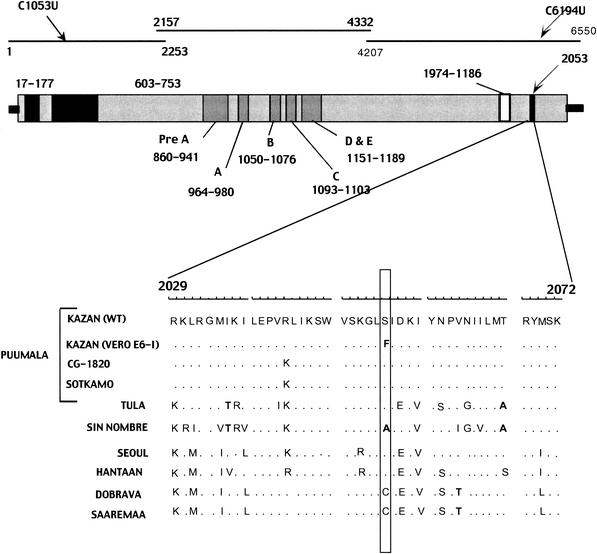
FIG. 1.
Schematic structure of hantaviral L segment. Black arrows indicate the locations of the differences found between the wild-type (WT) and Vero E6-I variants in the nucleotide and amino acid sequences. The three black lines indicate three fragments of the L segment that were amplified and sequenced separately, and numbers mark the position of each fragment in the nucleotide sequence. The gray bar below represents the L protein and its conserved regions, with numbers showing the location of each region in the amino acid sequence. The black stripe in the C terminus shows the site where the Ser2053Phe mutation resides. Alignment of hantaviral L protein sequences for the region surrounding the amino acid replacement Ser2053Phe is shown below, and nonhomologous amino acid substitutions observed in this region are shown in bold. Hantaviral sequences used for the alignment and retrieved from the GenBank include Hantaan virus strain 76-118 (X55901) (38), Seoul virus strain 80-39 (X56492) (2), Sin Nombre virus strain NM H10 (L37901) (4), Puumala virus strains CG1820 (M63194) (40) and Sotkamo (Z66548) (30), Tula virus strain Tula/Moravia/5302v (AJ005637) (23), Dobrava virus strain Ano-Poroia/9Af/99 (AJ410617) (29), and Saaremaa virus strain Saaremaa/160V (AJ410618) (29).