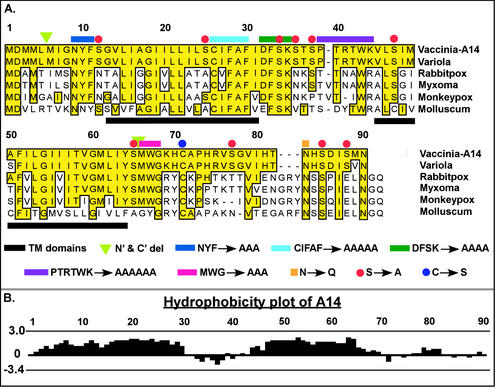
FIG. 1.
VV A14 is highly conserved among the Poxviridae family. (A) Alignment of VV A14 with several of its orthopox homologs. Putative membrane-spanning regions are underlined in black. The A14 mutants constructed for this study are depicted as follows: green triangles indicate the new initiation and termination sites engineered for alleles containing N′ and/or C′ truncations. Several sequences chosen for their high level of conservation among the Poxviridae homologs were changed to an equivalent number of Ala residues: N9YF (dark blue overline), C26IFAF (cyan), D32FSK (dark green), P37TRTWK (purple), and M66WG (magenta). Asn83 (orange square), part of a highly conserved N-linked glycosylation motif (NXS) at the C terminus of A14, was changed to Gln. Cys71 (blue circle), likely to direct the covalent dimerization of A14 dimer, was changed to Ser. Red circles highlight Ser residues at positions 12, 25, 34, 36, 38, 47, 65, 77, 85, and 88 that were mutated to Ala, individually and in various combinations (see text). (B) Hydrophobicity plot of VV A14 as determined by the Kyte-Doolittle method. The highly hydrophobic A14 protein is thought to span the membrane twice; the two transmembrane domains comprise residues 13 to 31 and 45 to 64, which are predicted to adopt inside-to-outside and outside-to-inside orientations, respectively (http://www.ch.embnet.org/software/TMPRED_form.html). The hydrophobic domains are separated by a 13-aa hydrophilic loop region.