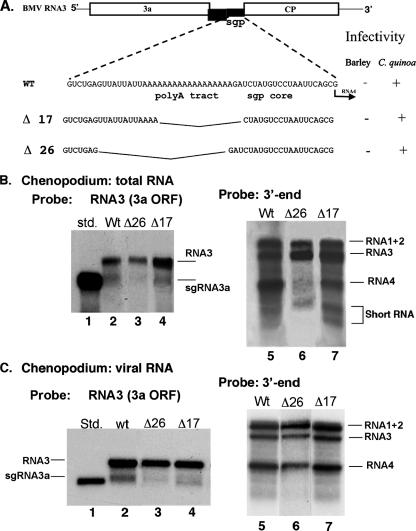
FIG. 4.
Effect of deletions in the oligo(A) tract on accumulation of sgRNA3a and sgRNA4 in vivo. (A) Schematic representation of the oligo(A) deletion constructs. The upper scheme shows the genomic RNA3 molecule and its sgp region sequence, while the lower part depicts deletion mutants. Δ26 lacks the entire oligo(A) sequence plus a short upstream region, and construct Δ17 carries only four A residues in the oligo(A) tract. (B) Northern blot analysis of accumulation of sgRNAs in total RNA extracts from Chenopodium quinoa. The RNA, extracted after 14 dpi as described in Materials and Methods, was separated in 1% denaturing agarose gel, transferred to a nylon membrane, and probed with an RNA3-specific probe (to detect sgRNA3a) or with a 3′-end probe (to detect sgRNA4). The inoculated RNA3 deletion constructs are indicated by the numbers on top, and the positions of individual RNAs are indicated on the right. (C) Northern blot analysis of accumulation of sgRNAs in the encapsidated BMV RNA from C. quinoa after 14 days psi, using two different probes, as specified. Std., in vitro transcribed 5′ 1,200 nt of RNA3.