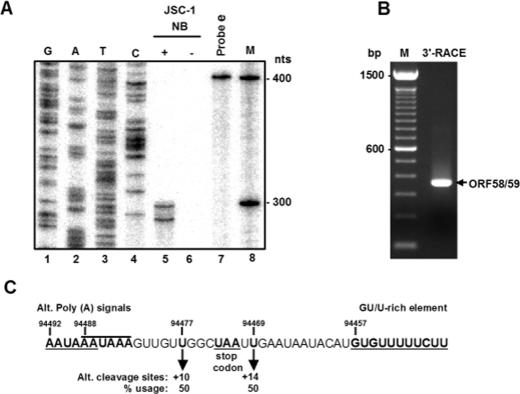
FIG. 5.
Mapping of the KSHV ORF58 and ORF59 polyadenylation cleavage sites by RPA and 3′ RACE. (A) RPA was performed on 20 μg of total RNA from uninduced or butyrate-induced (24 h) JSC-1 cells with 4 ng of 32P-labeled RNA probe e (see Fig. 4A). The protected RNA products were then compared with a sequence ladder that was generated by the antisense primer Pr94768 from a plasmid containing KSHV genomic DNA from nt 94368 to 94768. The protected products were separated along with the sequencing ladders and a 100-bp DNA ladder in an 8% denaturating polyacrylamide gel. (B) Mapping of ORF58 and ORF59 cleavage sites by 3′ RACE by using a primer, Pr94768 (see its position in Fig. 4A). An RACE product with the predicted size of ∼320 bp was detected, cloned, and sequenced. (C) The sequence reading showing two alternative cleavage sites from the mappings, along with two alternative overlapping poly(A) signals upstream and one GU/U-rich element downstream of the cleavage site. The +10 position is relative to the first poly(A) signal, and the +14 position is relative to the second.