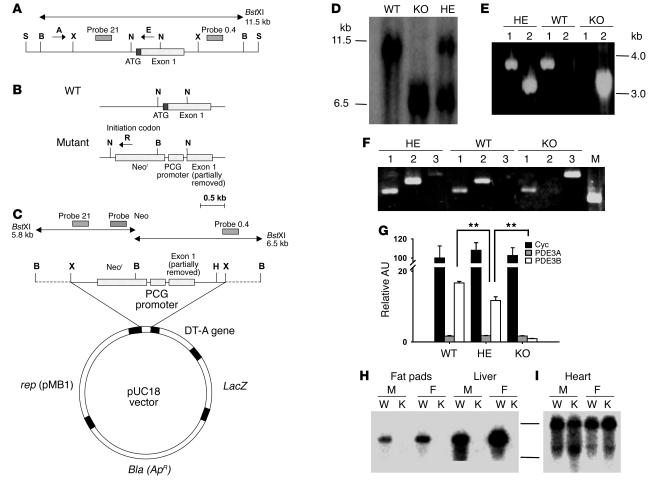
Figure 1. Targeted disruption of the murine Pde3b gene.
(A) Structure of the approximately 13-kb SalIPDE3B WT genomic fragment containing 5′-untranslated region and exon 1. (B) WT and disrupted KO gene fragments near exon 1. (C) Pde3b targeting vector. S, SalI; X, XbaI; B, BstXI; N, NotI; H, HindIII. (D) Southern blot of WT, HE, and KO genomic DNA, digested with BstXI and hybridized to 32P-labeled probe 0.4 (see A and C). (E) PCR amplification of WT, HE, and KO genomic DNA using the specific primers A, E, and R indicated in A and B (A and E for lane 1; A and R for lane 2). (F) RT-PCR amplification of mRNA from WT, HE, and KO livers with primers described in Methods targeted for PDE3A (lane 1), PDE3B (lane 2), and Neor (lane 3). M, molecular weight marker. (G) Quantitation of cyclophilin A (Cyc), PDE3A, and PDE3B mRNAs in WT, HE, and KO mouse livers by real-time RT-PCR. Data were normalized to the quantity of WT cyclophilin A mRNA, taken as 100 AU. Values represent mean ± SEM (n = 4 per genotype in triplicate assays). Data were similar in 2 other groups of WT and KO mice and 1 group of HE mice. **P < 0.01. (H and I) Northern blot of WT and KO mRNAs, using probe B for mPDE3B (fat pads and liver; H) or probe A for mPDE3A (heart; I) as described in Methods. M, male; F, female; W, WT mRNA; K, KO mRNA.