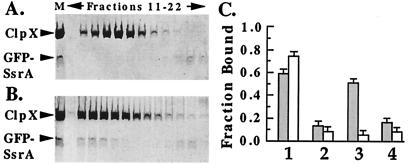
Figure 3.
Slow release of unfolded protein from ClpX in the presence of ATP[γS]. (A) ClpX (5 μM) was mixed with native GFP-SsrA (2.5 μM) in the presence of ATP[γS] and passed through a Superdex 200 gel filtration column (Amersham Pharmacia Biotech) in the presence of 1 mM ATP[γS]. Fractions (0.08 ml) were collected at 1-min intervals from 11 to 22 min, prepared for SDS/PAGE, separated on 10% polyacrylamide gels in SDS, and stained with Coomassie blue. Lane M, mixture before gel filtration. (B) ClpX (5 μM) was mixed with acid-denatured GFP-SsrA (2.5 μM) in the presence of ATP[γS], passed through Superdex 200, and analyzed as in A. (C) Ultrafiltration assays to compare ClpX binding of Gdn⋅HCl-denatured [3H]λO (gray columns) and folded native λO (white columns). ClpX (0.2 μM) was mixed with 0.1 μM λO in buffer with 2 mM ATP[γS] (column sets 1, 2, and 3) or 4 mM ATP (column set 4). The solutions were passed through a Microcon 100 ultrafiltration membrane (Amicon) by centrifugation for 5 min at 4°C, and the fraction of [3H]λO retained with ClpX was determined. Column set 1, binding of [3H]λO alone. Set 2, A 20-fold molar excess of unlabeled λO was added as a competitor at the same time as [3H]λO. Sets 3 and 4, after the [3H]λO had bound to ClpX for 5 min, the complex was incubated for 1 min with a 20-fold molar excess unlabeled λO before ultrafiltration.