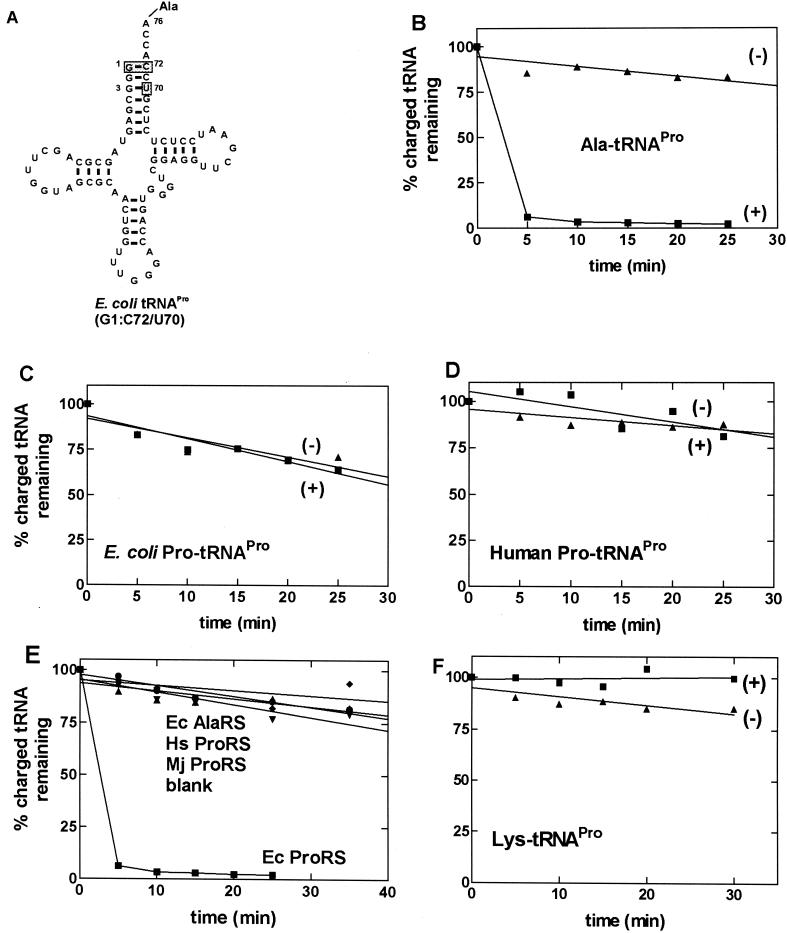
Figure 2.
Deacylation of wild-type tRNAPro and mischarged tRNAPro variants. (A) The sequence of alanine-accepting E. coli tRNAPro. The shaded boxes indicate nucleotides that are mutated relative to wild-type E. coli tRNAPro. (B–D) Graphs showing the deacylation of G1:C72/U70-[3H]Ala-tRNAPro (B), E. coli [3H]Pro-tRNAPro (C), and human [3H]Pro-tRNAPro (D) in the presence (+) and absence (−) of E. coli ProRS. (E) Graph showing that the efficient deacylation of the Ala-tRNAPro variant is specific for E. coli (Ec) ProRS (0.5 μM). E. coli AlaRS (1 μM) (⧫), human (Hs) ProRS (0.6 μM) (●), and M. jannaschii (Mj) ProRS (0.4 μM) (▾) were unable to deacylate E. coli G1:C72/U70-Ala-tRNAPro. (F) G1:C72/U70-[3H]Lys-tRNAPro is not deacylated by E. coli ProRS. In B-–F, the data obtained in the presence of E. coli ProRS are represented by ■, and the control reaction carried out in the absence of enzyme is represented by ▴.