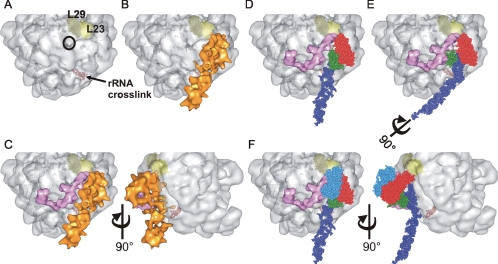
Figure 5.
Docking SRP on the ribosome. (A) Surface of the 50S ribosomal subunit from H. marismortui (PDB: 1W2B) highlighting subunits L23 (light yellow) and L29 (dark yellow) and the nascent chain exit site (black circle). The nucleotides which correspond to a region of 23S rRNA found to cross-link to bacterial SRP RNA are displayed as red spheres. (B) Alignment of the SRP reconstruction (orange) onto the 50S ribosomal subunit maximizing the overlap of regions of SRP with respective cross-linking sites on the ribosome (L23 and rRNA). (C) The structure of E. coli TF (pink) modeled onto the ribosome along with the SRP reconstruction. TF and SRP are in very close proximity to each other and flank the nascent chain exit site. A second view related to the first by rotation of 90° illustrates the effect of the kink in the RNA structure. (D) The SRP reconstruction was replaced with the ec_SRP model and demonstrates the orientation of M (green) with respect to the other domains of SRP, the large ribosomal subunit and TF. The SRP RNA, NG and M domains are colored as in Figure 3B. (E) Rotation of the SRP RNA by 90° moves the M domain of SRP in toward the nascent chain exit site where it can effectively compete with TF for sampling nascent chains as they emerge from the ribosome. (F) The structural data of the binding of the NG domain of the SRP receptor, FtsY (PDB: 1RJ9, blue), to E. coli Ffh NG was incorporated onto the ribosomal subunit. This location of FtsY is likely to result in the displacement of TF from the ribosome explaining mutually exclusive binding of TF and FtsY to ribosome–SRP complexes. Two views related by rotation of 90° are shown.