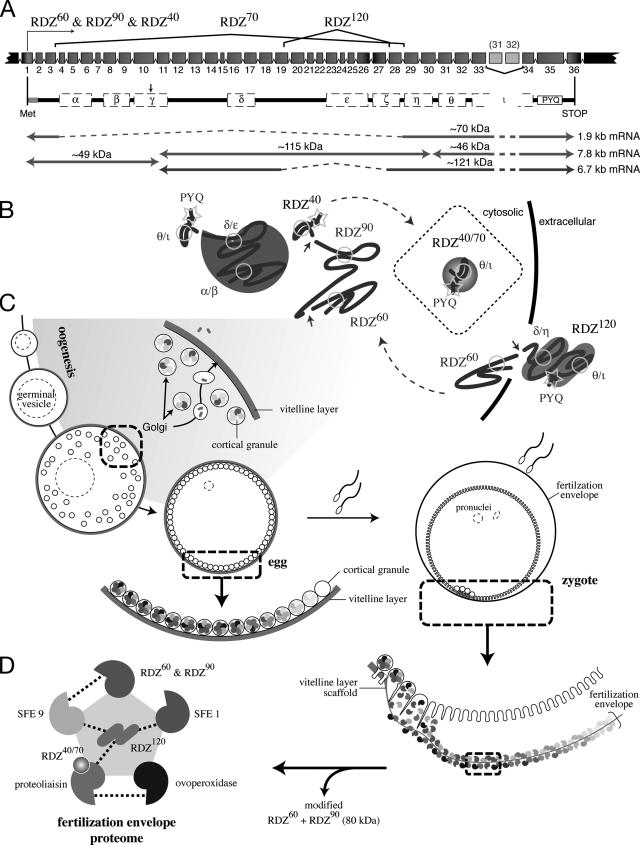
Figure 5.
Model of rendezvin protein expression and the fertilization envelope proteome. (A) Schematic of the S. purpuratus rendezvin exons, colored to match the predicted protein encoded, and resultant transcripts following posttranscriptional processing. The duplication of exons 31 (185 base pairs) and 32 (191 base pairs) is not retained in the most stable rdz transcript (8.2 kb versus stable 7.8 kb; our unpublished data). Spliced regions are shown with the corresponding isoform nomenclature used. Predicted primary structure, after removal of exons (dashed lines) and proteolysis (arrow; see text), are shown below with their predicted protein mass. The approximate transcript lengths that correspond to each isoform are listed on the right. Note the amino-terminal isoform (RDZ60; ∼49 kDa) is shared by two transcripts due to proteolysis. (B) Sketches of possible secondary structures for each RDZ component predicted by splice variants and immunoblot detection, including the interacting CUBs, are shown. (C) Diagram of oogenesis and fertilization, including a model for RDZ translation and protein trafficking within the oocyte (shaded region). Includes details of the egg and zygotic cortex undergoing exocytosis. (D) One binding network of interacting structural protein partners within the fertilization envelope is shown, following the color representation of Figure 4F.