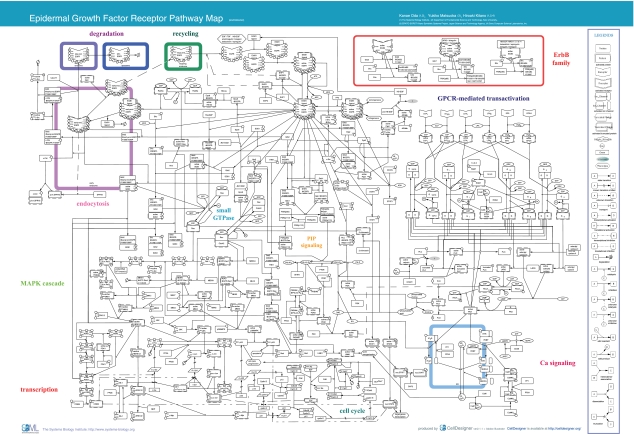
Figure 1.
EGFR Pathway Map. This map was created using CellDesigner ver. 2.0 (http://www.systems-biology.org/002/). A total of 219 reactions and 322 species were included. The map can be best viewed in the PDF format. Abi, abl-interactor; ADAM, a disintegrin and metalloproteinase; ADPR, ADP-ribose; Akt, v-akt murine thymoma viral oncogene homolog; AP-1, activator protein-1; Bad, BCL2-antagonist of cell death; cADPR, cyclic ADP-ribose; CAK, cyclin-dependent kinase-activating kinase; CaM, calmodulin; CaMK, calcium/calmodulin-dependent protein kinase; c-Cbl, Casitas B-lineage lymphoma proto-oncogene; CD, cluster of differentiation; Cdc, cell division cycle; Cdk, cyclin-dependent kinase; c-Fos, v-fos FBJ murine osteosarcoma viral oncogene; Chk, c-src tyrosine kinase (Csk) homologous kinase; c-Jun, v-jun sarcoma virus 17 oncogene homolog; c-Myc, v-myc myelocytomatosis viral oncogene homolog; CREB, cAMP response element-binding protein; c-Src, v-src sarcoma viral oncogene homolog; cyt., cytosol; DAG, diacylglycerol; EGF, epidermal growth factor; EGFR, epidermal growth factor receptor; Elk, Ets-like protein; end., endosome; EP, prostaglandin E receptor; Eps, EGF receptor pathway substrate; ER, endoplasmic reticulum; ErbB, erythroblastic leukemia viral (v-erb-b) oncogene homolog; ERK, extracellular signal-regulated kinase; Gab, GRB2-associated binding protein; GPCR, G protein-coupled receptor; Grb, growth factor receptor-bound protein; HB-EGF, heparin-binding EGF-like growth factor; IP3R, inositol 1,4,5-triphosphate receptor; IP3, inositol 1,4,5-triphosphate; JNK, c-Jun N-terminal kinase; KDI, kinase domain I; LARG, leukemia-associated rho guanine nucleotide exchange factor; LIMK, LIM (Lin-11 Isl-1 Mec-3) kinase; LPA, lysophosphatidic acid; LPA1/2, lysophosphatidic acid G protein-coupled receptor 1/2; lyso., lysosome; m., messenger; MAPK, mitogen-activated protein kinase; MEKK, MAPK/ERK kinase kinase; MKK, MAP kinase kinase; MKP, MAP kinase phosphatase; MLK, mixed lineage kinase; NAD, nicotinamide adenine dinucleotide; nuc., nucleus; PAK1, p21/Cdc42/Rac1-activated kinase; PDK, 3-phosphoinositide-dependent protein kinase; PGE2, prostaglandin E2; Pi, phosphoric ion; PI3,4,5-P3, phosphatidylinositol-3,4,5-triphosphate; PI3,4-P2, phosphatidylinositol-3,4-bisphosphate; PI3K, phosphatidylinositol-3-kinase; PI4,5-P2, phosphatidylinositol-4,5-bisphosphate; PI4-P, phosphatidylinositol-4-phosphate; PI5K, phosphatidylinositol-5-kinase; PIP, phosphatidylinositol polyphosphate; PKB, protein kinase B; PKC, protein kinase C; pl.m., plasma membrane; PLC, phospholipase C; PLD, phospholipase D; PP, protein phosphatase; PTB, phosphotyrosine-binding domain; PTEN, phosphatase and tensin homolog; Pyk, proline-rich tyrosine kinase; Rab5a, RAS-associated protein RAB5a; Raf, v-raf-1 murine leukemia viral oncogene homolog; Ras, rat sarcoma viral oncogene homolog; RasGAP, Ras GTPase-activating protein; Rb, retinoblastoma; RGS, regulator of G-protein signaling; Rin, Ras interaction; RN-tre, related to the N-terminus of tre; RSK, ribosomal protein S6 kinase; RYR, ryanodine receptor; S, serine; S1P, sphingosine-1-phosphate; S1P1/2/3, sphingolipid G protein-coupled receptor 1/2/3; SERCA, sarcoplasmic/endoplasmic reticulum calcium ATPase; Shc, Src homology 2 domain containing transforming protein; SHP, Shp-2 tyrosine phosphatase; SOS, son of sevenless homolog; SPRY, Sprouty; STAT, signal transducer and activator of transcription; T, threonine; TGFα, transforming growth factor alpha; Ubc, ubiquitin-conjugating enzyme; Y, tyrosine. This image is also available as high resolution PDF (see Supplementary PDF 1) or Scalable Vector Graphic (SVG) or SBML (see Supplementary SBML 1).