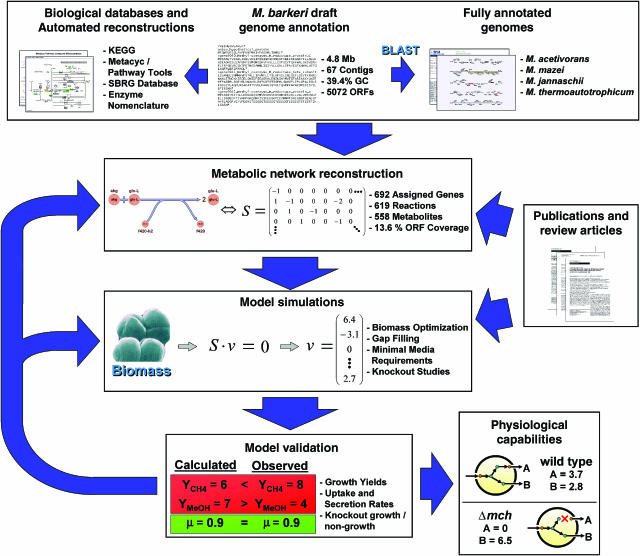
Figure 1.
The iterative model building procedure used to generate iAF692. The draft genome annotation was used as a scaffold, on which GPR assignments were made. The reactions added to the model were taken from both biochemical databases and published data. Once a reaction was found to be in the network, it was manually curated and either associated to a potential ORF or added with no gene assignment. A biomass objective function was formulated to perform model simulations based on cellular composition. Modeling simulations were run under steady-state conditions to determine the reaction flux distribution in the network. The results from the simulations were interpreted and compared to experimental data. From the comparison, physiological capabilities of the cell were confirmed or the network was further refined or updated.