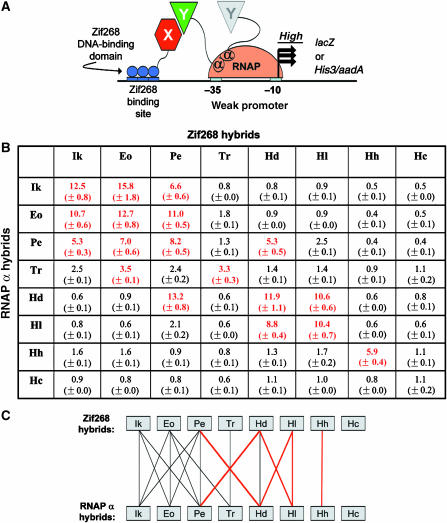
Figure 2.
Dimerization specificities of wild-type DZFs determined using a B2H system. (A) Schematic of B2H system used to study DZF interactions. Interaction of arbitrary protein domains X and Y fused to the Zif268 DBD and an E. coli RNAP α-subunit fragment, respectively, mediates recruitment of RNAP to a weak promoter bearing a Zif268 binding site. RNAP recruitment leads to increased transcription of an associated reporter gene (lacZ) or co-cistronic selectable marker genes (His3 and aadA). In the experiments of this report, domains X and Y are DZFs. Note that two α-subunits are present in an RNAP complex. (B) Pairwise combinations of eight wild-type DZF domains tested in the B2H system. Combinations of plasmids encoding wild-type DZFs were cotransformed into a B2H reporter strain harboring the lacZ gene and reporter gene expression was measured by β-galactosidase assay. Each combination was tested in triplicate with mean fold-activation values and standard errors of the means shown. Values highlighted in bold red text indicate positive interactions defined as >2.5-fold activation of LacZ expression. This cutoff was chosen based on the observation that the highest fold-activation observed in this assay for a noninteracting DZF pair (Ikaros and TRPS-1; McCarty et al, 2003) is 2.5. Ik=human Ikaros; Eo=human Eos; Pe=human Pegasus; Tr=human TRPS-1; Hd=Drosophila melanogaster Hunchback; Hl=Locusta migratoria Hunchback; Hh=Helobdella triserialis Hunchback; and Hc=Caenorhabditis elegans Hunchback. (C) Summary of DZF interaction specificity profiles determined using the B2H system. Lines indicate interactions detected among 64 pairwise combinations of eight wild-type DZFs (experiments of Figure 2B). Black lines indicate interactions that have also been detected by other methods and red lines indicate novel interactions that have not been described previously.