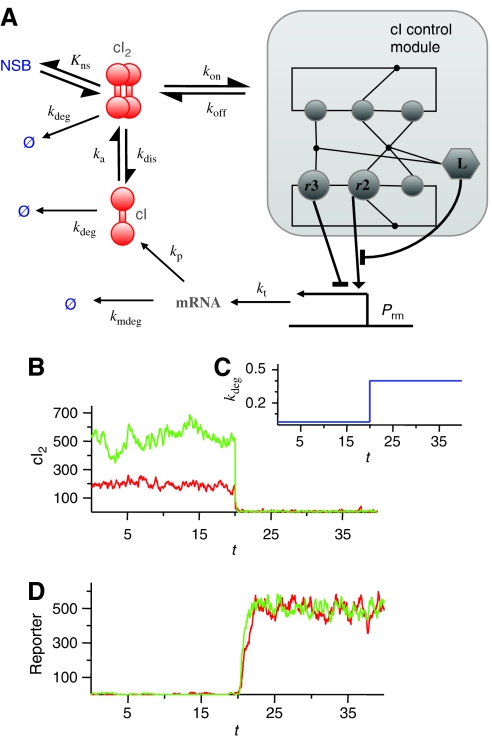
Figure 5.
Phage λ cI self-regulation kinetics. (A) The phage λ cI–DNA assembly network can we viewed as a module within the network that controls the production of cI repressors at the Prm promoter. Binding of cI dimers (cI2) to r3 (sr3=1), represses transcription, whereas binding to r2 when r3 is unoccupied (sr2=1 and sr3=0) activates transcription. The transcription rate is given by kt=(kactnl(1−sL)sr2+kactlsLsr2+kbas)(1−sr3), with kactnl=0.05 s−1, kactl=0.0225 s−1, and kbas=0.005 s−1; cI mRNA is degraded and translated into cI protein at rates kmdeg=0.005 s−1 and kp=0.05 s−1 per mRNA, respectively; and cI monomers and dimers are degraded at a rate kdeg. The cI dimerization reaction takes place with association and dissociation rate constants ka=7 × 108M−1s−1 and kdis=9.9 s−1, respectively. cI dimers in solution can bind nonspecifically to DNA, with an equilibrium binding constant Kns=2.0 × 104 M−1, or bind to one of the free operators with an association rate kon=a[N], with a=7 × 108M−1s−1. Note that the values used for the association rates are the typical ones of diffusion-limited reactions. The dissociation rates koff depend on the free energy difference between the complexes with (s) and without (s′) the cI dimer via the detailed balance principle: koff=kone−(ΔG(s′)−ΔG(s))/RT. As cellular volume, we have taken 10−15 l. The rates of looping and unlooping DNA are konL=30 s−1 (Vilar and Leibler, 2003) and koffL, respectively, with koffL obtained also from the detailed balance principle. (B) Time behavior of the number of cI dimers for the case with WT operators (red) and with no left operator (green) when (C) the cI degradation rate kdeg (in units of min−1) is switched from 0.025 min−1 to 0.4 min−1. Time on the horizontal axis is given in hours. (D) Activity of the promoter Pr controlling a reporter gene. In this case, there is transcription at a rate ktrep=0.12(1−sr1)(1−sr2)s−1 only when both r1 and r2 are free. The reporter mRNA translation, mRNA degradation, and protein degradation rates are kprep=0.01 s−1, kmdegrep=0.005 s−1, and kdegrep=0.005 s−1, respectively.