Networks of interacting genes are central to a cell's ability to sense and process information. Such networks are seldom comprised of simple linear cascades; feedback loops are ubiquitous and are implicated in processes such as cellular differentiation, circadian rhythm, and cell cycle control. Noise, or stochastic fluctuation in network components and the cellular environment, is an inherent feature of biochemical networks (Kaern et al, 2005). More importantly, fluctuations in a single molecular species can be transmitted via the network to affect multiple components. Given the abundance of feedback loops in such networks, it is not surprising that they can play important roles in noise processing. For instance, negative feedback loops can dampen fluctuations (Becskei and Serrano, 2000; Paulsson, 2004), whereas positive feedback loops can act as noise amplifiers that drive spontaneous state transitions (Figure 1A ). In an exciting paper by Süel et al (2006), two feedback loops coupled with noisy components in the competence induction network of Bacillus subtilis were shown to be responsible for its transient differentiation into the competent state.
Figure 1.
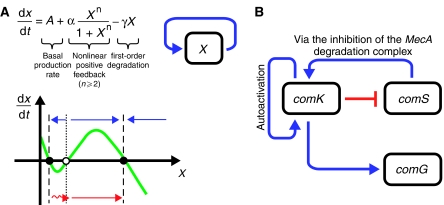
(A) Phase diagram illustrating how noise coupled with positive feedback can lead to spontaneous state transitions. The black dots denote stable fixed points and the blue arrows indicate the direction of flow when X is perturbed away from the fixed points. The red wiggling arrow denotes possible fluctuations away from one of the stable fixed points. Note that any fluctuation that goes past the unstable fixed point (white dot) can spontaneously drive the system toward the other state (in the case of the excitable competence induction network studied by Suel et al, fluctuations drive the system away from the one and only stable fixed point before eventually returning to the same fixed point). (B) A simplified version of the B. subtilis competence induction network. Note that comG is illustrated here to aid the description of the experiments; it is one of many proteins activated by comK, the master transcription factor in the process.
Natural genetic competence, or the ability to import DNA molecules from the environment, is a widespread phenomenon in bacteria. It facilitates horizontal gene transfer among bacterial species, and is likely important in DNA repair by taking in environmental DNA as templates (Solomon and Grossman, 1996). Not surprisingly, entrance into the competent state is regulated. In B. subtilis, most cells irreversibly sporulate in stationary phase when nutrients are limited, or when the cell density is high (Grossman, 1995). Interestingly, a small, but random population of cells enters the competent state transiently before returning to vegetative growth. This unique feature makes B. subtilis competence induction an ideal model to study the network architecture behind transient cellular differentiation.
At the core of the competence induction network is the master transcription factor ComK, which is at the center of a positive loop via self-activation and a negative feedback loop through ComS (Figure 1B). The stochastic nature of induction implicates intracellular noise as the potential driver of spontaneous differentiation. An intriguing possibility is that the underlying network is excitable such that fluctuations could drive ComK spontaneously to a high level (competence induction) via the positive feedback loop. High levels of ComK would in turn suppress the expression of ComS, leading to the eventual return of ComK to normal levels (competence exit). The time delay associated with the negative feedback determines the resident time in the competent state. Although this is an attractive hypothesis, the network topology is capable of other dynamic behaviors (see Supplementary information of Suel et al), and it is unclear what roles other genes in the network may play. So how does one find out whether the hypothesis is sound? The answer lies in the author's elegant single-cell measurements of network dynamics.
The authors start by inserting a pair of fluorescent proteins (YFP and CFP) driven by the comK and comG promoters into the B. subtilis chromosome, and observed that their expression in cells undergoing competence induction is highly correlated over time. Given that ComG is known to be primarily induced by ComK, the strong correlation implies that ComK is the key driver of its own expression during competence induction, thus confirming the positive feedback loop. To validate the negative feedback loop through ComS, the promoters of comS (PcomS) and comG (PcomG) were used to drive YFP and CFP expression, respectively. A strong negative correlation was observed between their expression dynamics: as the cell enters the competent state, CFP expression starts to go up whereas YFP expression goes down; the opposite was observed at competence exit. This strongly suggests that ComK negatively regulates ComS. More strikingly, the negative correlation was not observed in sister cells that are not undergoing competence induction, implying that the regulation is specific to the induction process.
The negative feedback through ComS basically implements a timer device to ensure that competent cells would eventually return to vegetative growth. To demonstrate that this feedback is important for the transient nature of the induction, the authors constructed a modified strain with an extra copy of comS driven by PcomG. If their model were correct, induced cells would fail to exit the competent state, as ComS would remain at a high level even when ComK is induced. This prediction was indeed observed and provided further proof of the authors' model. It also demonstrates the power of rewiring the underlying network in validating a strong quantitative prediction. However, it is still not certain whether the negative feedback is solely responsible for timely competence exit. One caveat of the above experiment is that the authors' modifications essentially introduce an extra positive feedback loop to ComK through ComS, hence induced cells are locked in the competent state as the network is now bi-stable (Figure 1A) instead of mono-stable.
This work opens up a host of interesting questions. For instance, it would be interesting to know the sources of the fluctuations. Do they mainly come from the intrinsic fluctuations of ComS/ComK or are other factors also involved? Perhaps tracking the fluctuation dynamics of ComS/ComK, both before and after the cells are stressed, would be a good start. Such measurements may also aid in the development of models that link levels of intracellular fluctuations to population statistics, such as the percentage of cells that undergo induction. It is also unclear whether and how levels of fluctuation are regulated—presumably factors that help dampen the fluctuation of ComS/ComK are needed to prevent premature inductions. Given the prevalence of competence induction in bacteria (Solomon and Grossman, 1996), it would be exciting to see whether similar network motifs operate in other species.
Recent works have highlighted the critical roles feedback loops play in establishing memory and maintaining stability in genetic networks (Xiong and Ferrell, 2003; Acar et al, 2005), as well as how fluctuations may drive phenotypic diversity in genetically identical populations (Kaern et al, 2005). The work of Süel et al (2006) further illustrates that such network motifs can be evolved to perform an important biological function. It is also encouraging that the main phenotypic feature of a complex network with a large number of components can be dissected by analyzing a few key players. Given that feedback loops with noisy parts can lead to a wide gamut of dynamical phenomenon, more surprises will surely be in store as we continue to explore how genetic networks link genotype to phenotype.
References
- Acar M, Becskei A, van Oudenaarden A (2005) Enhancement of cellular memory by reducing stochastic transitions. Nature 435: 228–232 [DOI] [PubMed] [Google Scholar]
- Becskei A, Serrano L (2000) Engineering stability in gene networks by autoregulation. Nature 405: 590–593 [DOI] [PubMed] [Google Scholar]
- Grossman AD (1995) Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu Rev Genet 29: 477–508 [DOI] [PubMed] [Google Scholar]
- Kaern M, Elston TC, Blake WJ, Collins JJ (2005) Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet 6: 451–464 [DOI] [PubMed] [Google Scholar]
- Paulsson J (2004) Summing up the noise in gene networks. Nature 427: 415–418 [DOI] [PubMed] [Google Scholar]
- Solomon JM, Grossman AD (1996) Who's competent and when: regulation of natural genetic competence in bacteria. Trends Genet 12: 150–155 [DOI] [PubMed] [Google Scholar]
- Süel G, Garcia-Ojalvo J, Liberman L, Elowitz MB (2006) An excitable gene regulatory circuit induces transient cellular differentiation. Nature 440: 545–550 [DOI] [PubMed] [Google Scholar]
- Xiong W, Ferrell JE Jr (2003) A positive-feedback-based bistable ‘memory module' that governs a cell fate decision. Nature 426: 460–465 [DOI] [PubMed] [Google Scholar]