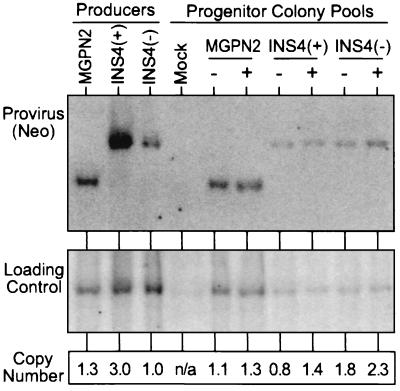
Figure 3.
Quantification of gene transfer rates by Southern blot analysis. DNA was isolated from the pools of progenitor colonies grown in the absence (−) and presence (+) of G418 selection used in the analysis presented in Fig. 2a, was digested with KpnI that cuts once in each LTR, and was analyzed by Southern blotting. The blot was first hybridized with a probe for Neo (Upper), was stripped, and was rehybridized with a loading control probe (mouse β-globin). The band intensities were quantified with a PhosphorImager and were used to calculate the relative number of provirus copies per genome in each sample (Lower). Controls included DNA from a pool of untransduced progenitor colonies (Mock), and DNA from producer clones for the three vectors.