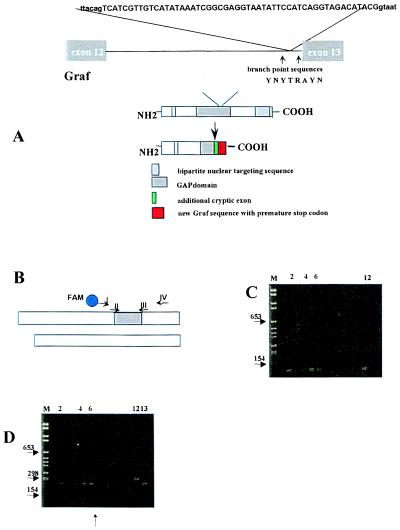
Figure 6.
(A) Insertion of 52-bp (capital letters) derived from intron 13 into the final cDNA found in patient #7. The surrounding intronic sequences are shown in lowercase letters. This leads to a reading frame shift followed by a premature stop codon. The GAP domain of Graf is substantially shortened. The intronic regions that were sequenced in patient #7 and 12 healthy controls are indicated by arrows. The splice branch site consensus sequence is shown as follows: Y represents T or C, R either A or G. (B) Schematic representation of both cDNA fragments that were coamplified by universal primers I and IV for assessment of their relative amount. Primers II and III amplify the aberrantly spliced fragment only. (C) Nested PCR analysis using the first-round primers I and IV and the second-round primers II and III. M, molecular weight marker. Lane 1, negative control. Four of 15 healthy blood donors expressed the aberrantly spliced fragment in their mononuclear cells (lanes 2 and 4–6) because a faint PCR product was seen. Lane 12, positive control. (D) Single-round PCR analysis using primers I and IV. Lane 6, two differently sized PCR products are seen from the cDNA of patient #7 even after only one round of PCR. Positive plasmid controls containing the 52-bp insertion (lane 12) or not (lane 13). In each RT-PCR, 2 μg of total RNA was subjected to cDNA synthesis and processed in parallel.