Figure 1.
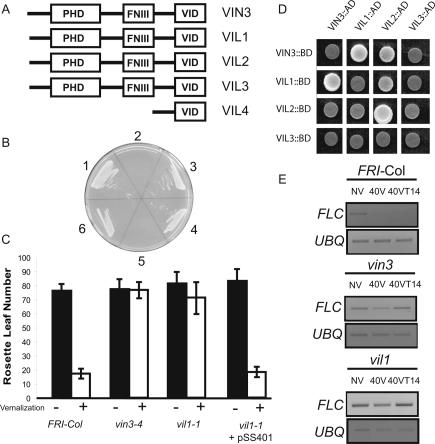
(A) Domain structures of VIN3-like proteins. (Amino acid alignments are provided in Supplementary Fig. 1.). VIN3 is At5g57380; VIL1 is At3g24440; VIL2 is At4g30200; VIL3 is At2g18880; and VIL4 is At2g18870. (B) The VID region is required for the interaction between VIN3 and VIL1. The constructs either have the VID region alone as described in Supplementary Figure S1 or the remainder of the gene without the VID region. Yeast two-hybrid assays: (1) full-length VIN3 as bait and full-length VIL1 as a prey. (2) VIN3 without VID as bait and full-length VIL1 as a prey. (3) VID of VIN3 as bait and full-length VIL1 as prey. (4) Full-length VIL1 as bait and full-length VIN3 as prey. (5) Full-length VIL1 as bait and VIN3 without VID as prey. (6) Full-length VIL1 as bait and VID of VIN3 as prey. Yeast two-hybrid assay conditions are described in Materials and Methods (Supplemental Material). (C) Yeast two-hybrid assays among the VID regions. (D) Vernalization response in vil1 and vin3. (Filled bars) Leaf number at flowering of nonvernalized plants; (open bars) values for plants cold-treated for 40 d as described in Materials and Methods (Supplemental Material). (E) FLC mRNA levels in vin3 and vil1. (NV) Samples from nonvernalized plants grown at 22°C; (40V) samples prepared directly from plants grown for 40 d at 4°C; (40VT14) plants grown for 40 d at 4°C followed by growth at 22°C for 14 d.