Figure 1.
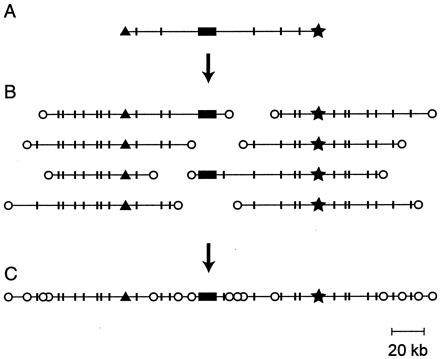
The virtual map. Recovery of a contig surrounding a known gene by use of the STC database. (A) The arrayed BAC library is screened by using a cDNA probe, and a BAC containing the gene sequence is selected; the most desirable clone would be the longest that has the gene sequence (solid box) toward the middle. The STCs for that BAC (▴, ★) are recovered from the database together with the length distribution of restriction fragments produced by the sites (indicated by short, vertical lines). (B) Oligonucleotide probes are created from the left and right STCs and used to scan the BAC library. The selected clones are aligned to overlap in the restriction digest patterns. The probes should not lie within repetitive sequences: if they do, too many BACs will score positive, and their restriction fragment lengths will not align. (C) The clones extending the longest distance left and right plus the original clone (A) constitute a contig surrounding the gene, marked by STC tags at frequent intervals.
