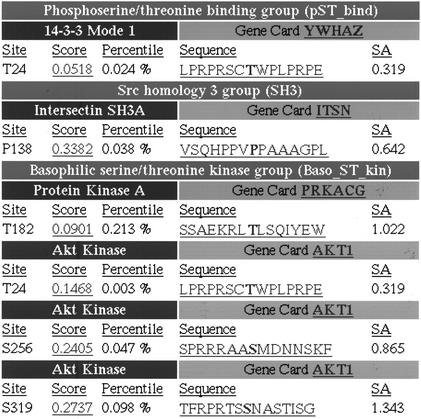
Figure 2.
Motif Scan's output table. For each motif family with a site on the graphical output (cf Fig. 1), details about the best matching domain motifs and the position of the site in the query are shown. The score, percentile and sequence of the site are indicated, as is the calculated surface accessibility for that site (labeled SA). Clicking on the score will display a histogram showing where this score ranks when compared with all potential sites for that motif in vertebrate SWISS-PROT; clicking on the sequence shows its position in the full protein and provides a link to BLAST for evaluating conservation of the motif in related protein homologues. For domains with an entry in the Weizman Institute's GeneCard database (http://bioinformatics.weizmann.ac.il/cards), the name is listed as a hyperlink to its GeneCard reference.